Arch Iran Med. 25(2):133-138.
doi: 10.34172/aim.2022.23
Mini Review
Prevention and Treatment of Hepatocellular Carcinoma Using miRNAs
Zahra Farzaneh 1, Maryam Farzaneh 2, 3, * 
Author information:
1Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
2Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
3Cellular and Molecular Research Center, Medical Basic Sciences Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
*Corresponding Author: Maryam Farzaneh, PhD; Fertility, Infertility and Perinatology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Email:
Maryamfarzaneh2013@yahoo.com
Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of death due to cancer. Liver transplantation, surgical liver resection, chemotherapy, and radiotherapy are the main options for the treatment of HCC. However, these methods are unable to limit the growth, survival, and metastasis of HCC cells. Several signaling pathways control propagation, metastasis, and recurrence of HCC. Recent studies have established new approaches for the prevention and treatment of HCC using miRNA technology. MicroRNAs are a class of non-coding RNAs with an average of 22 nucleotides that play critical roles in controlling gene expression in a variety of biological processes. miRNAs can induce or suppress HCC proliferation, migration, metastasis, and tumorigenesis. The anti-cancer effects of molecular agents can be evaluated directly in animal models or indirectly through the injection of HCC cell lines treated with anti-cancer agents. Targeting cancer-specific signaling pathways with miRNAs can be novel therapeutic strategies against HCC. This study provides the latest findings on using miRNAs in the control of HCC in both in vitro and in vivo models.
Keywords: Cancer, Hepatocellular carcinoma, miRNA, Signaling pathways
Copyright and License Information
© 2022 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article as: Farzaneh Z, Farzaneh M. Prevention and treatment of hepatocellular carcinoma using miRNAs. Arch Iran Med. 2022;25(2):133-138. doi: 10.34172/aim.2022.23
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death around the world.1-4 Non-viral (alcohol consumption and non-alcoholic fatty liver)5-7 and viral (hepatitis B/C virus) risk factors8,9 enhance the risk of HCC.10-13 There are three main options, including liver transplantation,14-16 surgical liver resection,17-19 and non-surgical methods (chemotherapy and radiotherapy) for the treatment of HCC.20-22 However, these approaches are unable to limit the progression and metastasis of HCC cells and cause side effects on the surrounding healthy cells.23,24 Several signaling pathways, including Wnt, Notch, EGF, SHH, hippo, and BMPs are associated with cell-division, metastasis, epithelial to mesenchymal transition (EMT), migration, and tumorigenesis of HCC.25-27 Targeting these signaling pathways may promote the treatment of the disease.28-30 Recent studies have established new approaches for the prevention and treatment of HCC using miRNA technology.31-33 microRNAs are a branch of RNA interference (RNAi) technology that contain about 20 nucleotides and target the specific mRNA in the cells.34,35 Evidence from miRNA expression profiles shows that some miRNAs are upregulated in HCC (oncomiR) and enhance the acquisition of metastatic potential.36,37 miRNAs can inhibit the expression of specific proteins (ligand or secondary messenger) in tumor-promoting signaling pathways and enhance HCC treatment efficacy.38,39 Molecularly targeted therapies using miRNAs with a high degree of specificity may be a suitable strategy in cancer treatment.31,40,41 This study provides the latest findings on using miRNAs in the control of HCC in both in vitro and in vivo models.
The Canonical miRNA Biogenesis
MicroRNAs are a class of non-coding RNAs with an average of 22 nucleotides that play an important role in controlling gene expression.42 miRNAs by microRNA-binding sites in the 3′ UTR of the target mRNAs trigger mRNA degradation to control the rate of translation.43 miRNAs can bind with the 5′ UTR, coding sequences, and gene promoters42 to regulate the expression of target genes or suppress translation by one of two distinct mechanisms.44 Pri-miRNAs or primary miRNAs are produced by the RNase II or III (poll III) in the nucleus.45,46 Subsequently, pri-miRNA with the Drosha/DGCR8 holoenzyme undergoes nuclear cleavage to produce a hairpin structured precursor or the precursor miRNA (pre-miRNA) with ∼60- to 70-nt.47 Exportin-5 (Exp5) and Ran-GTP can transport pre-miRNAs to the cytoplasm.48 Dicer is an RNase III endonuclease that combined with the transactivating response RNA-binding protein (TRBP) cleaves pre-miRNA hairpin to form a mature microRNA duplex (∼22-nt).49-51 Finally, miRNA binds with the AGO protein (RNA-induced silencing complex (RISC)) to target messenger RNA (mRNA) and stimulate mRNA cleavage, degradation, and translation repression.52,53 (Figure 1).
Targeting Signaling Pathways in HCC with miRNAs
Several previous studies have provided evidence that miRNA can suppress HCC metastasis54,55 (Table 1). miRNAs have been shown to control several signaling pathways, including Wnt, Notch, FGF, SHH, and hippo, and suppress the tumorigenesis of HCC (Figure 1).
Table 1.
Effects of miRNA on Signaling Pathways Related to HCC
|
Pathway
|
miRNA
|
Target
|
HCC cell line
|
Animal model
|
Result
|
Ref.
|
| TGF-B |
miR-200 |
Ligand |
MHCC-97 H, SMMC 7721, HepG2, Huh7 |
- |
Inhibit HCC proliferation, EMT, and invasion |
56
|
| miR-663 |
Ligand |
SK-Hep1, Huh7 and other HCC line |
- |
Inhibit the tumor growth and invasion |
57
|
| miR-34 |
Ligand |
Huh7, HepG2, Hep3B |
- |
Decrease the HCC proliferation |
58
|
| miR-133 |
|
SMMC7721, Huh7 |
1 × 107 SMMC7721 d subcutaneously to limb of nude mice |
Decrease proliferation, migration, increase apoptosis, decrease tumor growth |
59
|
| Wnt |
miR-298 |
B-catenin |
MHCC-97 H, HCCLM3 |
MHCC-97H subcutaneously to flank of nude mice |
Decrease the HCC proliferation and metastasis |
60
|
| miR-504 |
FZD receptor |
Huh7, HepG2 |
- |
Decrease the HCC proliferation and metastasis |
60
|
| miR-122 |
Pathway |
SMMC7721, Bel-7402 |
5×106 cells subcutaneously to flank of nude mice |
Decrease the HCC proliferation, survival and tumor weight |
61
|
| miR-148b |
Wnt1 |
HepG2 |
5 × 106 HepG2 subcutaneously to flank of nude mice |
Induce apoptosis and cell cycle arrest, inhibit tumor growth |
62
|
| miR- 3194-3p |
BCL9 |
MHCC-97H, Hep3B |
1 × 106 MHCC-97H or Hep3B to tail veins |
Inhibit migration, invasion, and metastasis |
63
|
| Shh |
miR-138 |
Smo receptor |
HepG2 |
- |
Decrease colony formation and invasion, increase apoptosis |
64
|
| miR-338-3p |
Smo receptor |
MHCC-97H |
1×107 MHCC-97H to flank of nude mice then cut and transplant to left liver |
Inhibit the EMT |
65
|
| Notch |
miR-3163 |
NICD |
MHCC97-H, LM-3 |
MHCC97-H subcutaneous or intraportal of nude mice |
Decrease the tumor growth |
66
|
| miR-206 |
NICD |
HepG2 |
- |
Cell cycle arrest, apoptosis, and inhibit the EMT |
67
|
| EGF |
miR-874 |
ERK |
SK-Hep1 |
overexpressed miR-874 SK-hep-1 to BALB/c nude mice |
Inhibit proliferation and metastasis, decrease the tumor size |
68
|
| HGF |
miR-181a-5p |
c-met |
SNU, Mahlavu |
- |
Inhibit proliferation and metastasis |
69
|
| VEGF |
miR-195 |
VEGF/FGF |
BEL-7402 |
- |
Inhibit migration and invasion |
70
|
| miR-126 |
EGF/VEGF |
HCCLM3, SMMC-7721, MHCC-97H |
subcutaneously to flank of SCID mouse |
Inhibit proliferation and tumor growth |
71
|
| Stat3 |
miR-345 |
Jak |
HCCLM3, HepG2 |
6×106 HCCLM3 cells intravenously into nude mice |
Inhibit the EMT and metastasis |
72
|
| miR-30e |
Jak |
HepG2, Huh7 |
- |
Inhibit the proliferation, migration, and invasion of HCC |
73
|
| YAP/TAZ |
miR-9-3p |
TAZ |
Huh1, HLF |
- |
Inhibit proliferation |
74
|
| miR186 |
YAP |
HepG2, Hep3B, SNU398 |
- |
Inhibit the migration and proliferation |
75
|
| HIF-1α |
miR-592 |
HIF-1α |
SK-hep1, SMMC-7721 |
1.5 × 106 SK-Hep-1 subcutaneously into flank of SCID mice |
Inhibit proliferation tumor growth, and glycolysis |
76
|
| Cell cycle |
miR-144 |
Cyclin B1 |
HepG2, SMMC-7721 |
5 × 106 SMMC-7721 subcutaneously to flanks of nude mice |
Decrease proliferation, migration, survival, and the tumor size |
77
|
| miR-214-3p |
Serin, theronin kinase |
HepG2, Huh7 |
- |
Decrease proliferation, increase the apoptosis |
56
|
| miR-300 |
B-Catenin |
HepG2, Huh7 |
- |
Decrease proliferation |
78
|
| Apoptosis |
miR-383 |
Stat3 |
HepG2, Huh7 |
DEN |
Increase the apoptosis, decrease proliferation |
79
|
| miR-644a |
Heat shock factor 1 |
HepG2, SMMC-7721 |
2 × 107 SMMC-7221 subcutaneously to nude mice |
Increase the apoptosis, decrease proliferation
inhibit tumor growth |
80
|
| miR-377 |
Bcl2 |
HepG2 |
- |
Inhibit proliferation and apoptosis |
81
|
| Autophagy |
miR-423-5p |
ATG7 |
Huh7 |
- |
Autophagy and cell cycle arrest |
82
|
| miR100 |
mTOR |
HepG2 |
5×106 HepG2 subcutaneously to flank of BALB/c mice |
Autophagy and apoptosis |
81
|
| ROS |
miR-125b-5p |
TXNRD |
Huh7, SK-hep1 |
- |
Decrease proliferation and migration |
81
|
| miR-124 |
SIRT1 |
Huh7, HepG2 |
5×106 HepG2 subcutaneously to armpit of nude mice |
Increase the apoptosis in combination with Cisplatin |
83
|
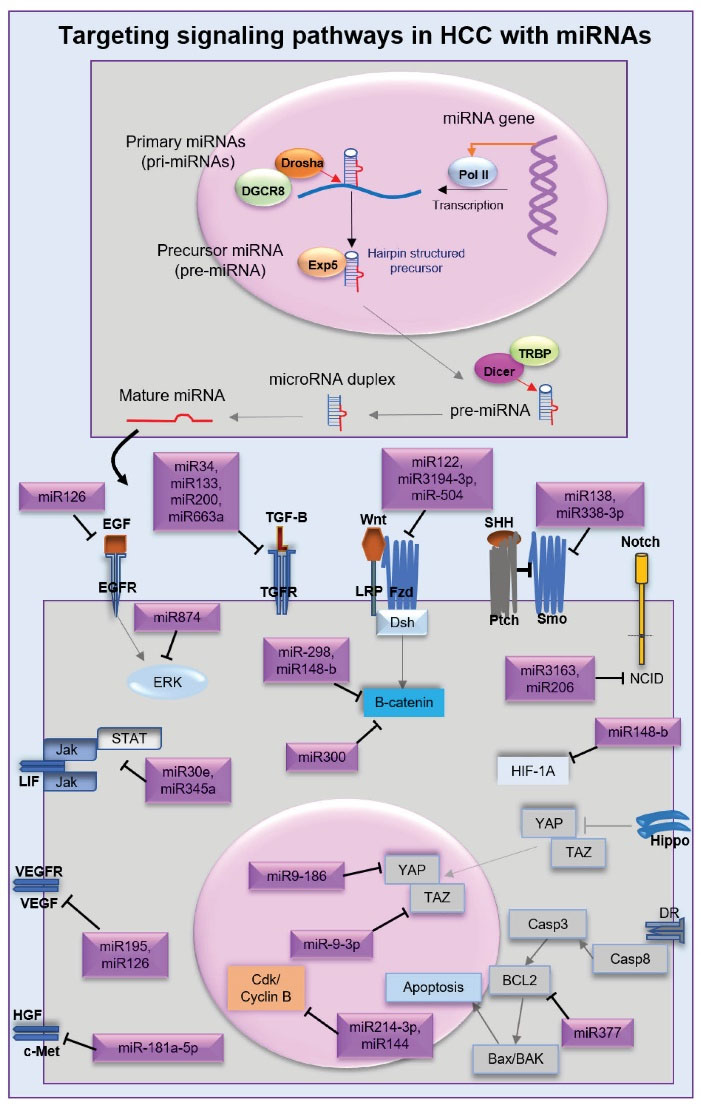
Figure 1.
miRNA Biogenesis and Anti-oncogenic Functions on Signaling Pathways Related to Hepatocellular Carcinoma.
.
miRNA Biogenesis and Anti-oncogenic Functions on Signaling Pathways Related to Hepatocellular Carcinoma.
It has been shown that overexpression of miR-34, miR-200, miR-133, and miR-663a inhibits the activation of the TGF-β ligand.56,57,59 miR-122 and miR-3194-3p have been found to suppress the Wnt/β-catenin pathway in HCC.61,63 Ectopic overexpression of miR-504 in HCC cells leads to blocking the FZD, while the overexpression of miR-298 and miR-148-b inhibits the activation of β-catenin.60,62 Overexpression of miR-13864 and miR-338-3p was shown to suppress Smo in the SHH pathway.65 Interestingly, SHH inhibitors accompanied by radiotherapy enhanced the radiosensitivity of HCC.84 miR-3163 and miR-206 have been reported to suppress the notch 1 intracellular domain (NICD) transcriptional activation in the Notch pathway.66,67 It has been confirmed that miR-874 blocks the EGF/ERK pathway in HCC.68 miR-181a-5p as a selective c-MET inhibitor in the HGF pathway decreases HCC proliferation, migration, and tumor growth.69,85-87 miR-195 inhibits angiogenesis by targeting VEGF and FGF,70 while miR-126 decreases the expression of VEGF and EGF.71 miR-30e and miR-345 are able to target the Jak/Stat3 pathway.72,73 miR-186 and miR-9-3p as tumor suppressors repress YAP75 and TAZ74 in the hippo pathway. Overexpression of miR-592 leads to disruption of hypoxia-inducible factor-1α (HIF-1α), suppression of glycolysis and lactate production, and reduction of G6PD mRNA levels in HCC.76
Anti-proliferative miRNA are significantly downregulated in HCC cell lines.78 Overexpression of miR-214-3p was reported that reduced HCC progression, by binding to the 3′-UTR of maternal embryonic leucine zipper kinase expression.88 miR-144 and miR-300 by targeting cyclin B and β-catenin, respectively, couldpromote cell cycle arrest in HCC.77,78,9 miR-383 by targeting IL-17 can suppress the Stat3 function, miR-644a inhibits heat shock factor 1 (an anti-apoptotic transcription factor), and miR-377 represses Bcl-2, thereby increasing apoptosis and decreasing cellular proliferation in HCC.79-81 Several studies have shown that miR100 and miR-423-5p induce autophagy.82,90 miR-124 interacts with sirtuin 1 (SIRT1) protein to enhance the cytotoxic effects of cisplatin in the CSC subpopulation.83
Taken together, targeting cancer-specific signaling pathways using miRNAs may be novel therapeutic strategies against HCC.
In conclusion, several important signaling pathways are misregulated in HCC compared to the normal hepatocytes.91,92 These pathways can trigger EMT, metastasis, migration, and tumorigenesis. Hence, suppression of the critical pathways with miRNAs causes cell cycle arrest, apoptosis, inhibits the tumorigenesis of HCC, and facilitates the sensitivity of HCC cells to drugs. Therefore, miRNAs may be a valuable approach to HCC treatment.93
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
Not applicable.
Conflict of Interest Disclosures
The authors declare no conflict of interest.
References
- Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: challenges and opportunities. Cancer Lett 2019; 460:1-9. doi: 10.1016/j.canlet.2019.114428 [Crossref] [ Google Scholar]
- Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 2016; 3:41-53. doi: 10.2147/jhc.s61146 [Crossref] [ Google Scholar]
- Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog 2017; 16:1. doi: 10.4103/jcar.JCar_9_16 [Crossref] [ Google Scholar]
- Forouzesh M, Hosseini M, Ataei M, Farzaneh M, Khoshnam SE. An extracellular matrix-based culture system for generation of human pluripotent stem cell derived-hepatocytes. Curr Stem Cell Res Ther 2021; 16(7):888-96. doi: 10.2174/1574888x16666201228144834 [Crossref] [ Google Scholar]
- Shen Y, Risch H, Lu L, Ma X, Irwin ML, Lim JK. Risk factors for hepatocellular carcinoma (HCC) in the northeast of the United States: results of a case-control study. Cancer Causes Control 2020; 31(4):321-32. doi: 10.1007/s10552-020-01277-1 [Crossref] [ Google Scholar]
- Scherübl H. Alcohol use and gastrointestinal cancer risk. Visc Med 2020; 36(3):175-81. doi: 10.1159/000507232 [Crossref] [ Google Scholar]
- Smeuninx B, Boslem E, Febbraio MA. Current and future treatments in the fight against non-alcoholic fatty liver disease. Cancers (Basel) 2020; 12(7):1714. doi: 10.3390/cancers12071714 [Crossref] [ Google Scholar]
- D’Souza S, Lau KC, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J Gastroenterol 2020; 26(38):5759-83. doi: 10.3748/wjg.v26.i38.5759 [Crossref] [ Google Scholar]
- Hiraoka A, Nagamatsu K, Izumoto H, Adachi T, Yoshino T, Tsuruta M. Zinc deficiency as an independent prognostic factor for patients with early hepatocellular carcinoma due to hepatitis virus. Hepatol Res 2020; 50(1):92-100. doi: 10.1111/hepr.13430 [Crossref] [ Google Scholar]
- Iida-Ueno A, Enomoto M, Tamori A, Kawada N. Hepatitis B virus infection and alcohol consumption. World J Gastroenterol 2017; 23(15):2651-9. doi: 10.3748/wjg.v23.i15.2651 [Crossref] [ Google Scholar]
- Midorikawa Y, Takayama T, Nakayama H, Higaki T, Moriguchi M, Moriya K. Prior hepatitis B virus infection as a co-factor of chronic hepatitis C patient survival after resection of hepatocellular carcinoma. BMC Gastroenterol 2019; 19(1):147. doi: 10.1186/s12876-019-1069-y [Crossref] [ Google Scholar]
- Li W, Deng R, Liu S, Wang K, Sun J. Hepatitis B virus-related hepatocellular carcinoma in the era of antiviral therapy: the emerging role of non-viral risk factors. Liver Int 2020; 40(10):2316-25. doi: 10.1111/liv.14607 [Crossref] [ Google Scholar]
- Chu YJ, Yang HI, Wu HC, Lee MH, Liu J, Wang LY. Aflatoxin B1 exposure increases the risk of hepatocellular carcinoma associated with hepatitis C virus infection or alcohol consumption. Eur J Cancer 2018; 94:37-46. doi: 10.1016/j.ejca.2018.02.010 [Crossref] [ Google Scholar]
- Goumard C, Scatton O. Resectable HCC: Should salvage liver transplantation for HCC be discussed de principe?. Clin Res Hepatol Gastroenterol 2020; 44(2):117-8. doi: 10.1016/j.clinre.2019.11.001 [Crossref] [ Google Scholar]
- Lee DD, Sapisochin G, Mehta N, Gorgen A, Musto KR, Hajda H. Surveillance for HCC after liver transplantation: increased monitoring may yield aggressive treatment options and improved postrecurrence survival. Transplantation 2020; 104(10):2105-12. doi: 10.1097/tp.0000000000003117 [Crossref] [ Google Scholar]
- Habibollahi P, Sheth RA, Cressman ENK. Histological correlation for radiofrequency and microwave ablation in the local control of hepatocellular carcinoma (HCC) before liver transplantation: a comprehensive review. Cancers (Basel) 2020; 13(1):104. doi: 10.3390/cancers13010104 [Crossref] [ Google Scholar]
- Chen KY, Huang YH, Teo WH, Chang CW, Chen YS, Yeh YC. Loss of Tid1/DNAJA3 co-chaperone promotes progression and recurrence of hepatocellular carcinoma after surgical resection: a novel model to stratify risk of recurrence. Cancers (Basel) 2021; 13(1):138. doi: 10.3390/cancers13010138 [Crossref] [ Google Scholar]
- Liu Q, Li J, Zhou L, Gu H, Wu K, You N. Liver parenchyma transection-first approach for laparoscopic left hemihepatectomy: a propensity score matching analysis. World J Surg 2021; 45(2):615-23. doi: 10.1007/s00268-020-05846-y [Crossref] [ Google Scholar]
- Midorikawa Y, Takayama T, Nakayama H, Moriguchi M, Aramaki O, Yamazaki S. Favorable outcomes of surgical resection for extrahepatic recurrent hepatocellular carcinoma. Hepatol Res 2020; 50(8):978-84. doi: 10.1111/hepr.13526 [Crossref] [ Google Scholar]
- Lee JS, Chon YE, Kim BK, Park JY, Kim DY, Ahn SH. Prognostic value of alpha-fetoprotein in patients who achieve a complete response to transarterial chemoembolization for hepatocellular carcinoma. Yonsei Med J 2021; 62(1):12-20. doi: 10.3349/ymj.2021.62.1.12 [Crossref] [ Google Scholar]
- Lee HA, Park S, Seo YS, Yoon WS, Shin IS, Rim CH. Surgery versus external beam radiotherapy for hepatocellular carcinoma involving the inferior vena cava or right atrium: a systematic review and meta-analysis. J Hepatobiliary Pancreat Sci 2021; 28(12):1031-46. doi: 10.1002/jhbp.865 [Crossref] [ Google Scholar]
- Yuan S, Guo Y. Hepatocellular Carcinoma with Right Atrial Tumor Thrombus: A Systematic Review. Research Square 2020. doi: 10.21203/rs.2.22554/v2 [Crossref]
- Lin YL, Li Y. Study on the hepatocellular carcinoma model with metastasis. Genes Dis 2020; 7(3):336-50. doi: 10.1016/j.gendis.2019.12.008 [Crossref] [ Google Scholar]
- Orcutt ST, Anaya DA. Liver resection and surgical strategies for management of primary liver cancer. Cancer Control 2018; 25(1):1073274817744621. doi: 10.1177/1073274817744621 [Crossref] [ Google Scholar]
- Swamy SG, Kameshwar VH, Shubha PB, Looi CY, Shanmugam MK, Arfuso F. Targeting multiple oncogenic pathways for the treatment of hepatocellular carcinoma. Target Oncol 2017; 12(1):1-10. doi: 10.1007/s11523-016-0452-7 [Crossref] [ Google Scholar]
- Alqahtani A, Khan Z, Alloghbi A, Said Ahmed TS, Ashraf M, Hammouda DM. Hepatocellular carcinoma: molecular mechanisms and targeted therapies. Medicina (Kaunas) 2019; 55(9):526. doi: 10.3390/medicina55090526 [Crossref] [ Google Scholar]
- Chatterjee S, Sil PC. Targeting the crosstalks of Wnt pathway with Hedgehog and Notch for cancer therapy. Pharmacol Res 2019; 142:251-61. doi: 10.1016/j.phrs.2019.02.027 [Crossref] [ Google Scholar]
- Lachenmayer A, Alsinet C, Chang CY, Llovet JM. Molecular approaches to treatment of hepatocellular carcinoma. Dig Liver Dis 2010; 42 Suppl 3:S264-72. doi: 10.1016/s1590-8658(10)60515-4 [Crossref] [ Google Scholar]
- Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel) 2020; 12(2):491. doi: 10.3390/cancers12020491 [Crossref] [ Google Scholar]
- Farzaneh Z, Vosough M, Agarwal T, Farzaneh M. Critical signaling pathways governing hepatocellular carcinoma behavior; small molecule-based approaches. Cancer Cell Int 2021; 21(1):208. doi: 10.1186/s12935-021-01924-w [Crossref] [ Google Scholar]
- Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci 2019; 110(4):1140-7. doi: 10.1111/cas.13965 [Crossref] [ Google Scholar]
- Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in hepatitis B virus-related hepatocellular carcinoma: potential as biomarkers and therapeutic targets. Front Oncol 2020; 10:1271. doi: 10.3389/fonc.2020.01271 [Crossref] [ Google Scholar]
- Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res 2018; 37(1):324. doi: 10.1186/s13046-018-0965-2 [Crossref] [ Google Scholar]
- Farzaneh M, Alishahi M, Derakhshan Z, Sarani NH, Attari F, Khoshnam SE. The expression and functional roles of miRNAs in embryonic and lineage-specific stem cells. Curr Stem Cell Res Ther 2019; 14(3):278-89. doi: 10.2174/1574888x14666190123162402 [Crossref] [ Google Scholar]
- Xu W, Jiang X, Huang L. RNA interference technology. In: Moo-Young M, Cui Z, Ye H, eds. Comprehensive Biotechnology. 3rd ed. Elsevier; 2019. p. 560-75. 10.1016/b978-0-444-64046-8.00282-2.
- Zhao WT, Lin XL, Liu Y, Han LX, Li J, Lin TY. miR-26a promotes hepatocellular carcinoma invasion and metastasis by inhibiting PTEN and inhibits cell growth by repressing EZH2. Lab Invest 2019; 99(10):1484-500. doi: 10.1038/s41374-019-0270-5 [Crossref] [ Google Scholar]
- Yang C, Dou R, Yin T, Ding J. MiRNA-106b-5p in human cancers: diverse functions and promising biomarker. Biomed Pharmacother 2020; 127:110211. doi: 10.1016/j.biopha.2020.110211 [Crossref] [ Google Scholar]
- Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Masaki T. MicroRNAs in the pathogenesis of hepatocellular carcinoma: A review. Cancers 2021; 13:514. [ Google Scholar]
- He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S. miRNA-based biomarkers, therapies, and resistance in cancer. Int J Biol Sci 2020; 16(14):2628-47. doi: 10.7150/ijbs.47203 [Crossref] [ Google Scholar]
- Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R. Circulating microRNAs in cancer: potential and challenge. Front Genet 2019; 10:626. doi: 10.3389/fgene.2019.00626 [Crossref] [ Google Scholar]
- Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem 2014; 6(17):1967-84. doi: 10.4155/fmc.14.116 [Crossref] [ Google Scholar]
- O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018; 9:402. doi: 10.3389/fendo.2018.00402 [Crossref] [ Google Scholar]
- Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014; 2014:970607. doi: 10.1155/2014/970607 [Crossref] [ Google Scholar]
- Vaschetto LM. miRNA activation is an endogenous gene expression pathway. RNA Biol 2018; 15(6):826-8. doi: 10.1080/15476286.2018.1451722 [Crossref] [ Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006; 13(12):1097-101. doi: 10.1038/nsmb1167 [Crossref] [ Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005; 123(4):631-40. doi: 10.1016/j.cell.2005.10.022 [Crossref] [ Google Scholar]
- Zeng C, Xia J, Chen X, Zhou Y, Peng M, Zhang W. MicroRNA-like RNAs from the same miRNA precursors play a role in cassava chilling responses. Sci Rep 2017; 7(1):17135. doi: 10.1038/s41598-017-16861-w [Crossref] [ Google Scholar]
- Wu K, He J, Pu W, Peng Y. The role of exportin-5 in microRNA biogenesis and cancer. Genomics Proteomics Bioinformatics 2018; 16(2):120-6. doi: 10.1016/j.gpb.2017.09.004 [Crossref] [ Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15(8):509-24. doi: 10.1038/nrm3838 [Crossref] [ Google Scholar]
- Graves P, Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics 2012; 10(5):239-45. doi: 10.1016/j.gpb.2012.06.004 [Crossref] [ Google Scholar]
- Fareh M, Yeom KH, Haagsma AC, Chauhan S, Heo I, Joo C. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat Commun 2016; 7:13694. doi: 10.1038/ncomms13694 [Crossref] [ Google Scholar]
- Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci 2019; 76(3):441-51. doi: 10.1007/s00018-018-2940-7 [Crossref] [ Google Scholar]
- Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 2015; 4(9):e252. doi: 10.1038/mtna.2015.23 [Crossref] [ Google Scholar]
- Hung CH, Chiu YC, Chen CH, Hu TH. MicroRNAs in hepatocellular carcinoma: carcinogenesis, progression, and therapeutic target. Biomed Res Int 2014; 2014:486407. doi: 10.1155/2014/486407 [Crossref] [ Google Scholar]
- Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z. The role of microRNAs in hepatocellular carcinoma. J Cancer 2018; 9(19):3557-69. doi: 10.7150/jca.26350 [Crossref] [ Google Scholar]
- Chen SY, Ma DN, Chen QD, Zhang JJ, Tian YR, Wang ZC. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer 2017; 8(4):617-25. doi: 10.7150/jca.17394 [Crossref] [ Google Scholar]
- Zhang C, Chen B, Jiao A, Li F, Sun N, Zhang G. miR-663a inhibits tumor growth and invasion by regulating TGF-β1 in hepatocellular carcinoma. BMC Cancer 2018; 18(1):1179. doi: 10.1186/s12885-018-5016-z [Crossref] [ Google Scholar]
- Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang Y. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res 2014; 74(21):6236-47. doi: 10.1158/0008-5472.can-14-0855 [Crossref] [ Google Scholar]
- Sun L, Guo Z, Sun J, Li J, Dong Z, Zhang Y. MiR-133a acts as an anti-oncogene in hepatocellular carcinoma by inhibiting FOSL2 through TGF-β/Smad3 signaling pathway. Biomed Pharmacother 2018; 107:168-76. doi: 10.1016/j.biopha.2018.07.151 [Crossref] [ Google Scholar]
- Cao N, Mu L, Yang W, Liu L, Liang L, Zhang H. MicroRNA-298 represses hepatocellular carcinoma progression by inhibiting CTNND1-mediated Wnt/β-catenin signaling. Biomed Pharmacother 2018; 106:483-90. doi: 10.1016/j.biopha.2018.06.135 [Crossref] [ Google Scholar]
- Cao F, Yin LX. miR-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting MDR1 by targeting Wnt/β-catenin pathway. Exp Mol Pathol 2019; 106:34-43. doi: 10.1016/j.yexmp.2018.10.009 [Crossref] [ Google Scholar]
- Zhang JG, Shi Y, Hong DF, Song M, Huang D, Wang CY. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/β-catenin pathway. Sci Rep 2015; 5:8087. doi: 10.1038/srep08087 [Crossref] [ Google Scholar]
- Yao B, Li Y, Wang L, Chen T, Niu Y, Liu Q. MicroRNA-3194-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by decreasing Wnt/β-catenin signaling through targeting BCL9. Artif Cells Nanomed Biotechnol 2019; 47(1):3885-95. doi: 10.1080/21691401.2019.1670190 [Crossref] [ Google Scholar]
- Luo J, Chen P, Xie W, Wu F. MicroRNA-138 inhibits cell proliferation in hepatocellular carcinoma by targeting Sirt1. Oncol Rep 2017; 38(2):1067-74. doi: 10.3892/or.2017.5782 [Crossref] [ Google Scholar]
- Chen JS, Liang LL, Xu HX, Chen F, Shen SL, Chen W. miR-338-3p inhibits epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma cells. Oncotarget 2017; 8(42):71418-29. doi: 10.18632/oncotarget.10138 [Crossref] [ Google Scholar]
- Yang B, Wang C, Xie H, Wang Y, Huang J, Rong Y. MicroRNA-3163 targets ADAM-17 and enhances the sensitivity of hepatocellular carcinoma cells to molecular targeted agents. Cell Death Dis 2019; 10(10):784. doi: 10.1038/s41419-019-2023-1 [Crossref] [ Google Scholar]
- Liu W, Xu C, Wan H, Liu C, Wen C, Lu H. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med 2014; 34(2):420-8. doi: 10.3892/ijmm.2014.1800 [Crossref] [ Google Scholar]
- Zhang Y, Wei Y, Li X, Liang X, Wang L, Song J. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis 2018; 9(2):130. doi: 10.1038/s41419-017-0131-3 [Crossref] [ Google Scholar]
- Korhan P, Erdal E, Atabey N. MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-Met. Biochem Biophys Res Commun 2014; 450(4):1304-12. doi: 10.1016/j.bbrc.2014.06.142 [Crossref] [ Google Scholar]
- Wang M, Zhang J, Tong L, Ma X, Qiu X. MiR-195 is a key negative regulator of hepatocellular carcinoma metastasis by targeting FGF2 and VEGFA. Int J Clin Exp Pathol 2015; 8(11):14110-20. [ Google Scholar]
- Hu MH, Ma CY, Wang XM, Ye CD, Zhang GX, Chen L. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget 2016; 7(41):66922-34. doi: 10.18632/oncotarget.11877 [Crossref] [ Google Scholar]
- Yu M, Xue H, Wang Y, Shen Q, Jiang Q, Zhang X. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int J Oncol 2017; 50(3):975-83. doi: 10.3892/ijo.2017.3852 [Crossref] [ Google Scholar]
- Mao J, Hu X, Pang P, Zhou B, Li D, Shan H. miR-30e acts as a tumor suppressor in hepatocellular carcinoma partly via JAK1/STAT3 pathway. Oncol Rep 2017; 38(1):393-401. doi: 10.3892/or.2017.5683 [Crossref] [ Google Scholar]
- Higashi T, Hayashi H, Ishimoto T, Takeyama H, Kaida T, Arima K. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br J Cancer 2015; 113(2):252-8. doi: 10.1038/bjc.2015.170 [Crossref] [ Google Scholar]
- Ruan T, He X, Yu J, Hang Z. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett 2016; 11(4):2941-5. doi: 10.3892/ol.2016.4312 [Crossref] [ Google Scholar]
- Jia YY, Zhao JY, Li BL, Gao K, Song Y, Liu MY. miR-592/WSB1/HIF-1α axis inhibits glycolytic metabolism to decrease hepatocellular carcinoma growth. Oncotarget 2016; 7(23):35257-69. doi: 10.18632/oncotarget.9135 [Crossref] [ Google Scholar]
- Gu J, Liu X, Li J, He Y. MicroRNA-144 inhibits cell proliferation, migration and invasion in human hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int 2019; 19:15. doi: 10.1186/s12935-019-0729-x [Crossref] [ Google Scholar]
- Bai J, Gao Y, Du Y, Yang X, Zhang X. MicroRNA-300 inhibits the growth of hepatocellular carcinoma cells by downregulating CREPT/Wnt/β-catenin signaling. Oncol Lett 2019; 18(4):3743-53. doi: 10.3892/ol.2019.10712 [Crossref] [ Google Scholar]
- Wang J, Lu L, Luo Z, Li W, Lu Y, Tang Q. miR-383 inhibits cell growth and promotes cell apoptosis in hepatocellular carcinoma by targeting IL-17 via STAT3 signaling pathway. Biomed Pharmacother 2019; 120:109551. doi: 10.1016/j.biopha.2019.109551 [Crossref] [ Google Scholar]
- Liang W, Liao Y, Li Z, Wang Y, Zheng S, Xu X. MicroRNA-644a promotes apoptosis of hepatocellular carcinoma cells by downregulating the expression of heat shock factor 1. Cell Commun Signal 2018; 16(1):30. doi: 10.1186/s12964-018-0244-z [Crossref] [ Google Scholar]
- Ge H, Zou D, Wang Y, Jiang H, Wang L. MicroRNA-377 downregulates Bcl-xL and increases apoptosis in hepatocellular carcinoma cells. Oncol Res 2017; 25(1):29-34. doi: 10.3727/096504016x14719078133168 [Crossref] [ Google Scholar]
- Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids 2015; 4:e233. doi: 10.1038/mtna.2015.8 [Crossref] [ Google Scholar]
- Xu Y, Lai Y, Weng H, Tan L, Li Y, Chen G. MiR-124 sensitizes cisplatin-induced cytotoxicity against CD133+ hepatocellular carcinoma cells by targeting SIRT1/ROS/JNK pathway. Aging (Albany NY) 2019; 11(9):2551-64. doi: 10.18632/aging.101876 [Crossref] [ Google Scholar]
- Della Corte CM, Viscardi G, Papaccio F, Esposito G, Martini G, Ciardiello D. Implication of the Hedgehog pathway in hepatocellular carcinoma. World J Gastroenterol 2017; 23(24):4330-40. doi: 10.3748/wjg.v23.i24.4330 [Crossref] [ Google Scholar]
- You H, Ding W, Dang H, Jiang Y, Rountree CB. c-Met represents a potential therapeutic target for personalized treatment in hepatocellular carcinoma. Hepatology 2011; 54(3):879-89. doi: 10.1002/hep.24450 [Crossref] [ Google Scholar]
- Luo T, Zhang SG, Zhu LF, Zhang FX, Li W, Zhao K. A selective c-Met and Trks inhibitor Indo5 suppresses hepatocellular carcinoma growth. J Exp Clin Cancer Res 2019; 38(1):130. doi: 10.1186/s13046-019-1104-4 [Crossref] [ Google Scholar]
- Du Z, Caenepeel S, Shen Y, Rex K, Zhang Y, He Y. Preclinical evaluation of AMG 337, a highly selective small molecule MET inhibitor, in hepatocellular carcinoma. Mol Cancer Ther 2016; 15(6):1227-37. doi: 10.1158/1535-7163.mct-15-0745 [Crossref] [ Google Scholar]
- Li Y, Li Y, Chen Y, Xie Q, Dong N, Gao Y. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int 2017; 17:102. doi: 10.1186/s12935-017-0471-1 [Crossref] [ Google Scholar]
- Semaan L, Zeng Q, Lu Y, Zhang Y, Zreik MM, Chamseddine MB. MicroRNA-214 enriched exosomes from human cerebral endothelial cells (hCEC) sensitize hepatocellular carcinoma to anti-cancer drugs. Oncotarget 2021; 12:185. [ Google Scholar]
- Ge YY, Shi Q, Zheng ZY, Gong J, Zeng C, Yang J. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget 2014; 5(15):6218-28. doi: 10.18632/oncotarget.2189 [Crossref] [ Google Scholar]
- Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J, Kim H. Hippo signaling interactions with Wnt/β-catenin and Notch signaling repress liver tumorigenesis. J Clin Invest 2017; 127(1):137-52. doi: 10.1172/jci88486 [Crossref] [ Google Scholar]
- Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther 2020; 5(1):8. doi: 10.1038/s41392-020-0110-5 [Crossref] [ Google Scholar]
- Abdel-Hamid NM, Abass SA, Mohamed AA, Muneam Hamid D. Herbal management of hepatocellular carcinoma through cutting the pathways of the common risk factors. Biomed Pharmacother 2018; 107:1246-58. doi: 10.1016/j.biopha.2018.08.104 [Crossref] [ Google Scholar]