Arch Iran Med. 27(2):62-71.
doi: 10.34172/aim.2024.11
Original Article
Gut-Lung Microbiota Characterization in Patients with Non-Small Cell Lung Carcinoma and COVID-19 Coinfection
Bahareh Vakili Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, 1 
Parisa Shoaei Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing – original draft, 2, * 
Kiana Shahzamani Formal analysis, Writing – review & editing, 3 
Seyed Davar Siadat Conceptualization, Methodology, 4, 5 
Hasan Shojaei Data curation, Writing – review & editing, 6 
Zahra Esfandiari Data curation, Writing – review & editing, 7 
Elahe Nasri Supervision, 1 
Shiva Shabani Supervision, 8 
Ali Zamani Moghadam Investigation, 1
Behrooz Ataei Data curation, Supervision, 1 
Author information:
1Infectious Diseases and Tropical Medicine Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
2Nosocomial Infection Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
3Hepatitis Research Center, School of Medicine, Lorestan University of Medical Sciences, Khoramabad, Iran
4Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran, Tehran, Iran
5Microbiology Research Center (MRC), Pasteur Institute of Iran, Tehran, Iran
6Department of Microbiology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
7Department of Food Science and Technology, Nutrition and Food Security Research Center, School of Nutrition and Food Science, Isfahan University of Medical Sciences, Isfahan, Iran
8Department of Infectious Diseases, School of Medicine, Arak University of Medical Sciences, Arak, Iran
Abstract
Background:
Non-small cell lung cancer (NSCLC) patients with COVID-19 have an excessive chance of morbidity and mortality. The fecal-nasopharyngeal microbiota compositions of NSCLC patients were assessed in this study.
Methods:
In total, 234 samples were collected from 17 NSCLC patients infected with COVID-19, 20 NSCLC patients without confirmed COVID-19, 40 non NSCLC patients with COVID-19, and 40 healthy individuals.
Results:
In lung microbiota, the abundance of Streptococcus spp. in NSCLC patients with confirmed COVID-19 was significantly higher than the two control groups. Pseudomonas aeruginosa and Staphylococcus aureus were listed as the most frequent pulmonary bacterial groups that colonized COVID-19 patients. In fecal specimens, the numbers of Bacteroidetes, Firmicutes, and Actinobacteria phyla were significantly higher amongst NSCLC patients with COVID-19. NSCLC patients infected with COVID-19 showed lower levels of Lactobacillus spp., Akkermansia muciniphila, and Bifidobacterium spp. The counts of Streptococcus spp., in NSCLC patients with COVID-19 were significantly higher than those of healthy individuals (8.49±0.70 log CFU/g wet feces vs 8.49±0.70 log CFU/g wet feces). Prevotella spp. were enriched in the gut and respiratory tracts of COVID-19 patient groups. The unbiased analysis showed an increment in Enterococcus spp., Streptococcus spp., and Prevotella spp.
Conclusion:
Eventually, it was found that compared to control groups, COVID-19 patients with NSCLC showed diminished gut bacteria diversity and increase in Lactobacillus spp., A. muciniphila, and Bifidobacterium spp. The overgrowth of Enterococcus spp., Streptococcus spp., and Prevotella spp. could be potential predictive biomarkers in the gut-lung axis of NSCLC patients with COVID-19.
Keywords: COVID-19, Gut-lung microbiota, Molecular characterization, Non-small cell lung cancer, Real-time PCR
Copyright and License Information
© 2024 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article as: Vakili B, Shoaei P, Shahzamani K, Siadat SD, Shojaei H, Esfandiari Z, et al. Gut-lung microbiota characterization in patients with non-small cell lung carcinoma and COVID-19 coinfection. Arch Iran Med. 2024;27(2):62-71. doi: 10.34172/ aim.2024.11
Introduction
The coronavirus infectious disease 2019 (COVID-19) resulted in a pandemic with a broad range of respiratory and gastrointestinal manifestations which contributed to more than 3.7 million global deaths.1 Trillions of symbiotic microbial cells inhabit human body parts, notably the skin, gastrointestinal (GI) tract and respiratory tract. Dysbiosis disrupts immune homeostasis because of an imbalance in microflora.2 Gut dysbiosis is associated with different GI and non-GI comorbidities, like inflammatory bowel disease, colorectal cancer, diabetes, hypertension, or obesity.3-5 The incidence of digestive symptoms in COVID-19 cases is reported from 2% to 50%; consequently, the GI tract is responsible for disease severity, and viral transmission.6
The dominant gut microbial population of healthy adults incorporates the phyla Firmicutes, Bacteroidetes, and predominant genera include Enterococcus, Faecalibacterium, Bacteroides, and Prevotella.1 In the microbial communities of healthy lungs, the dominant species including Bacteroidetes, Firmicutes, Proteobacteria, and the prevailing genera incorporate Streptococcus, Fusobacterium, Pseudomonas, Veillonella, and Prevotella.7,8 Balanced gut microbiota affects our health notably, but dysbiosis of the gut microbial population results in pathobiont proliferation and reduced commensal bacteria that cause different local and long-lasting diseases.1,5,7
Lung diseases impact the gut microbiota; in this way, a gut-lung axis permits the movement of endotoxins, cytokines, and microbial metabolites into the blood circulation and links the gut niche with that of one of the lungs.5,9
In COVID-19 patients with diarrhea, the gut-lung microbiota axis involves the circulatory and immune systems.10,11 The immune system signals modulate bacterial patterns particularly and increment the number of Escherichia coli or Pseudomonadales.12
COVID-19 patients with cancer are at great risk of serious morbidity and mortality.13 The diversity of gut microbiota is mainly reduced in elderly patients because of the leaking of SARS-CoV-2, hence COVID-19 has been more severe in this vulnerable groups by enabling SARS-CoV-2 to leak into the bloodstream and reach the internal organs.4 Although bacterial metabolites and fragments can modulate the lung immunity, altered gut and lung microbiota in COVID-19 patients can adjust the risk of developing severe respiratory complications, secondary bacterial infections, and death in those patients.14-16 However, data specifically focused on the effects of respiratory microbiota on the gut, the composition of lung microbiota in COVID-19 patients with lung cancer, and the principal risk factors for lung cancer patients with COVID-19 are scarce.14,17 About 1.6 million lung cancer patients succumb each year to the disease, of which non-small cell lung cancer (NSCLC) is the prevalent type of lung cancer histology (induced by acquired environmental effects and host genetic factors).17 Lung cancer describes a unique condition of cumulative risks for COVID-19 problems, such as advanced age, numerous comorbidities, prolonged smoking, lung tumors, and immunosuppressive treatment.13,18 Subsequently, lung cancer patients may be more defenseless against viral infections with poor prognosis if COVID-19 is suspected.13 Several studies from China, the United States, and Europe have detailed wide ranges of coinfection, from 2% to 80%.19 We utilized quantitative real-time polymerase chain reaction (qRT-PCR) in detecting targeted respiratory and gut bacteria; however, the use of next-generation sequencing (NGS) has low heterogeneity in the detection methods and could reveal coinfections that may be missed. We conducted a case-control study on NSCLC patients. The study included four groups of participants: 1st group; NSCLC patients, 2nd group; NSCLC patients infected with COVID-19 and 3th Group; COVID-19 patients and 4th group; healthy individuals.
The principle objective of the current study was to investigate the gut and lung microbiota of NSCLC cancer patients and whether or not the presence of SARS-CoV-2 could alter their resident microbiota.
Materials and Methods
Study Design and Base Line Characteristics
This case-control research was performed over 12 months at the referral cancer hospital of Isfahan University of Medical Sciences, Isfahan, Iran. It is a specialized care hospital for the treatment of hematology-oncology patients. This well-equipped 166-bed center has provided specialized medical care to the community for over 50 years
This study included 17 NSCLC patients infected with COVID-19, 20 NSCLC patients without COVID-19 infection, 40 COVID-19 patients without cancer, and 40 healthy persons who were enrolled from the beginning of April 2020 to the end of March 2021 (1:2 matched positives NSCLC to negative NSCLC). COVID-19 patients without cancer and healthy individuals were randomly selected; in addition, their sex and age were matched to NSCLC patients infected with COVID-19.
Inclusion and Exclusion Criteria for Patients
All hospitalized NSCLC patients were included in the study, regardless of when cancer had been identified and only based on the persistence of SARS-CoV-2 and a certain clinical consequence (discharge or death). The survey was carried out before the start of general vaccination against COVID-19 epidemics and the participants did not receive any dose of the COVID-19 vaccines.
NSCLC patients with evidence of surgery or neoadjuvant chemotherapy were included.NSCLC patients with acute digestive infection in the past two months who required antibiotic treatment or with a history of previous intestinal cancer treated by radiotherapy or surgery were not included. NSCLC patients with previous oropharyngeal, gastrectomy, or patients who were moved to other hospitals were also excluded.
Healthy control individuals were chosen from people with negative results of real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for SARS-CoV-2 tests (Rotor-Gene Q 6000, Qiagen, Hilden, Germany) who were referred to the reference laboratories of Isfahan. COVID-19 patients without cancer and healthy control subjects were without any disorder, pregnancy, chemotherapeutic or antimicrobial therapy for more than 3 months prior to sampling. Healthy individuals were defined as never smokers, without any respiratory diseases proved by normal chest computed tomography (CT) images or chronic disorders, within one year. CT scans were evaluated due to the presence of disorders such as ground-glass opacities in the lung. A total of 234 specimens, including 117 nasal swabs, and 117 feces samples, were gathered from the patients by professionals at the hospital. The nasopharynx and fecal samples were directly placed into sterile containers and then immediately transported to the laboratory of Infectious Diseases and Tropical Medicine Research Center, Isfahan, Iran, and stored long-term at − 80 °C. These samples were then utilized for profiling the gut and nasopharyngeal microbiota in all patients and healthy controls. The study was limited to individuals who met the specified inclusion criteria to reduce the effects of potential confounders including a food frequency questionnaire (FFQ).
Data Collection
Laboratory data (including hemoglobin, white blood cells, neutrophils, lymphocytes, platelets, albumin, creatinine, D-dimer, C-reactive protein, procalcitonin) were all collected at the time of admission (Tables S1 and S2). Data on comorbidities (diabetes, hypertension, cardiovascular disease), radiographic and pathologic findings, CT scans and lung cancer status, were collected from medical records (Tables 1 and 2).
Table 1.
Comparison of Clinical Characteristics of NSCLC Patients with and without COVID-19
|
Variables
|
NSCLC Patients with COVID-19 (n=17)
|
NSCLC Patients without COVID-19 (n=20)
|
P
Value
|
| History of smoking |
15 (88.2) |
18(90) |
0.63 |
| Comorbidities |
|
|
|
| Hypertension |
5 (29.4) |
7 (35) |
0.49 |
| Diabetes |
1 (5.9) |
2 (10) |
|
| Cardiovascular disease |
3 (17.6) |
5 (25) |
0.44 |
| Others (kidney, liver) |
- |
2 (10) |
0.28 |
| Clinical stage* |
|
|
|
| I |
7 (41.2) |
14 (70) |
0.25 |
| II |
1 (5.9) |
- |
| III |
3 (17.6) |
3 (15) |
| IV |
6 (35.3) |
3 (15) |
| Previous hospitalization within 2 months |
7 (41.2) |
4 (20) |
0.15 |
| History surgery within 3 months |
5 (29.4) |
3 (15) |
0.25 |
| History of pneumonitis |
4 (23.5) |
3 (15) |
0.41 |
| Use of immunomodulatory drug within 2 months |
4 (23.5) |
6 (30) |
0.47 |
| Use of corticosteroids within 2 months |
6 (35.3) |
5 (25) |
0.37 |
| Chemotherapy treatment within 4 weeks before admission |
11(64.7) |
6 (30) |
0.04 |
n/N (%) the number of patients with available data. Chi-square test for differences between two groups.
*Stage groups for NSCLC. Stage I: a small tumor that has not spread to any lymph nodes, Stage II: tumors (size:4-5 cm) can be removed with surgery, but often additional treatments are recommended, Stage III: have often spread extensively to the lymph nodes, but have not spread to other distant parts of the body, Stage IV: the lung cancer has spread to more than 1 area in the other lung, heart or distant parts of the body through the bloodstream.
Table 2.
Comparison of Clinical Characteristics of NSCLC Patients and COVID-19 Patients
|
Characteristics
|
NSCLC Patient with COVID-19 (n=17)
|
COVID-19 Patients (n=40)
|
P
Value
|
| Clinical presentation |
|
|
|
| Fever |
13 (76.47) |
26 (65) |
0.30 |
| Cough |
14 (82.35) |
19 (47.5) |
0.01 |
| Dyspnea (respiratory symptoms) |
9 (52.94) |
11 (27.5) |
0.06 |
| Vomiting |
3 (17.64) |
10 (25) |
0.41 |
| GI symptoms (Diarrhea) |
3 (17.64) |
5 (12.5) |
0.45 |
| Median spo2 % |
83 (87-80) |
91 (94-85) |
0.04 |
| Ground-glass opacity |
14 (82.35) |
22 (55) |
0.04 |
| Hospitalization |
|
|
|
| Admission to ICU |
9 (52.9%) |
9 (22.5) |
0.03 |
| Oxygen therapy |
|
|
|
| Need for supplement O2 |
10 (58.82) |
18 (45) |
0.25 |
| Invasive mechanical ventilation |
7 (41.17) |
10 (25) |
0.04 |
| Pneumonia- Non-sever |
10 (58.82) |
31 (77.5) |
0.13 |
| COVID-19 severity |
|
|
|
| Mild |
1 (5.88) |
8 (20) |
0.19 |
| Moderate |
7 (41.17) |
23 (57.5) |
| Sever |
9 (52.9) |
9 (22.5) |
| Drug administration of COVID-19 |
| Antibiotics |
15 (88.23) |
21 (52.5) |
0.01 |
| Antiviral |
14 (82.35) |
12 (30) |
< 0.001 |
| Median number of antibiotics administered |
5 (3-6) |
2 (1-3) |
0.05 |
| Median number of days exposed to treatment antibiotics (duration of antibiotic exposure) |
16 (5-23) |
6 (3-15) |
0.04 |
| Median duration of hospital stay |
18 (5-31) |
9 (4-21) |
0.04 |
n/N (%) the number of patients with available data. Chi- square test for differences between two groups.
Definitions
Diagnosis of COVID-19 was based on WHO’s temporary guideline.20 COVID-19 pneumonia was confirmed by clinical criteria and laboratory nucleic acid detection confirmed on genes targeted N and ORF1ab.21
COVID-19 patients were categorized in three groups, moderate: patients with respiratory symptoms of pneumonia; severe: patients with shortness of breath, and venous oxygen saturation < 93% in resting state; and critical: hospitalized patients with acute respiratory distress.
Colonization was defined as the presence of microorganisms, including detection of the bacteria (threshold of 102 CFU/mL). Above this threshold, some microbial flora discerns potential pathogens in immunocompetent.22
The quantities of respiratory pathogens including Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenza, Pseudomonas aeruginosa, and Moraxella catarrhalis in COVID-19 patients groups were taken into account. Moreover, other bacterial agents belonging to the gastrointestinal or nasopharyngeal flora such asBacteroidetes, Firmicutes, Actinobacteria phyla, Prevotella spp., Veillonella spp., Streptococcus spp., Enterococcus spp., Enterobacteriaceae, Lactobacillus spp., Clostridium spp., Bifidobacterium spp., Fusobacterium spp., Faecalibacterium prausnitzii, and Akkermansia muciniphila were considered. Nasopharyngeal swabs and stool samples were examined for the qRT-PCR based on different targeted bacteria quantities as described previously.23
Total DNA Isolation
Total DNA extraction from 500 microliters of nasopharyngeal swabs and 220 mg of stool was performed using QIAmp DNA stool and Blood mini kits (Qiagen) following the manufacturer’s instructions.24 DNAs were purified and stored at –80 °C for further experiments. DNA concentration was quantified using the QubitTM 4Florometer (Life Tech, Invitrogen, Singapore) before performing qRT-PCR.
Quantitative Real-Time PCR Analysis
The SYBR Green PCR Master Mix (Applied Biosystems) in Rotor-Gene 6000 real-time PCR cycler (Qiagen Corbett, Hilden, Germany), using the unique QuantiTect SYBR® Green PCR kit qPCR master mix (Yekta Tajhiz Azma Co, Tehran, Iran) and the proper qPCR primers, determined the bacterial load based on the abundance of 16S rRNA gene (Supplementary file 1, Table S3).24-29
About 50 ng of DNA from fecal and nasopharyngeal swabs (containing targeted species DNA), 1 × SYBR green qPCR master mix, and 0.5 μM of each primer were applied in each qPCR assay to generate standard curves for the enumeration of target DNA in test samples. All tests were assessed in duplicate, and the mean values were evaluated, as well. Every run contained non-template and positive controls. Amplification was performed under the following conditions: 95 °C for 5 minutes, followed by 40 cycles of denaturation at 95 °C for 1 minute, 30 s at the appropriate annealing temperature and 72 °C for 1 minute, and the final step extension at 72 °C for 5 minutes. The melting curve analysis was performed after qPCR with constant fluorescence inspection under a gradual heating rate of 0.1 °C/s from 72 to 95 °C. Standard curves were generated for assessment of the number of bacterial strains in samples with serially diluted solutions of the total extracted DNA from the reference strains. This method is reliable and consistent, and its accuracy is appropriate.
The bacterial concentrations (from fecal and nasopharyngeal swabs) were calculated by competitive cycle threshold values and presented as DNA copy numbers in reactions. Table S4 supplies standard curve parameters such as strains of bacteria, qPCR amplification efficiency, and determination coefficient. The correlation coefficient values of the standard curve ranged from 0.99 to 1.0.
Statistical Analysis
The SPSS software version 21.0 (SPSS Inc. Chicago, IL, USA) was performed for statistical calculations. Values were estimated as mean ± standard deviation (SD). The Shapiro–Wilk test was applied for testing the normality. The Q-Q plot was also performed for assumption of the normality. One-way analysis of variance (ANOVA) and Pearson’s chi-square were used for continuous variables (e.g. body mass index, age, and inter-patient comparisons) and categorical variables, respectively. A P value < 0.05 was considered statistically significant. We performed a post hoc test to identify exactly the differences between groups. ANOVA (Bonferroni’s, Dunnett’s tests) was performed in multiple comparisons between the groups. Box-plot graphs showed the median values of qPCR results as lines across the box. A t-test was performed to compare the means of the two groups of COVID-19 patients without cancer and healthy control subjects.
Results
A total of 234 samples were evaluated including 117 nasal swabs, and 117 feces samples from the patients and the healthy control group (Figure 1). The clinical characteristics and basic information of studied patients are shown in Tables 1 and 2. The mean age of NSCLC patients with COVID-19 was 58.7 (range 53.3-64.1), and 82.3% of them were male. They represented considerably higher coughs and received more antibiotics and antiviral agents compared to NSCLC patients without COVID-19 (P < 0.01, P < 0.01, and P 0.001, respectively).
Figure 1.
Flow Diagram Summarizing the Information of Samples and Patients in This Study
.
Flow Diagram Summarizing the Information of Samples and Patients in This Study
No significant difference was found in terms of BMI, age, or COVID-19 severity between patient groups and the control group (Table S5). Results revealed that the rate of chemotherapy treatment 4 weeks before admission in COVID-19 patients with NSCLC was significantly more than the NSCLC patients with negative COVID-19 (P = 0.04). CT images in NSCLC patients with COVID-19 showed more ground-glass opacity than patients with COVID-19 (P = 0.04). FFQ analysis showed no significant differences between the studied groups when they were compared for dietary intake of proteins and carbohydrates and fat.
Quantitation of Lung Microbiota
A comparison of quantities of selected microbial groups in lung microbiota in studied patients and healthy people is shown in Figure 2 and Table 3 (Table S6). Table 3 shows the average counts and the standard deviation of lung-targeted microorganisms. It showed that the plenitude of Streptococcus spp. in NSCLC patients suffering from COVID-19 was significantly higher than healthy subjects and NSCLC patients (157 ± 49.01; P < 0.003, 126 ± 34.71; P < 0.001, and141 ± 47.56; P < 0.002, respectively) (Table 3 and Figure 2). An expanded quantity of Prevotella spp. was found in COVID-19 patients with and without NSCLC (P < 0.05). P. aeruginosa and S. aureus were listed as the most frequent pulmonary bacterial groups colonizing in COVID-19 patients (Table 4). Furthermore, P. aeruginosa colonization was revealed in the lung microbiota of 35.3% (6/17) of NSCLC patients suffering from COVID-19.
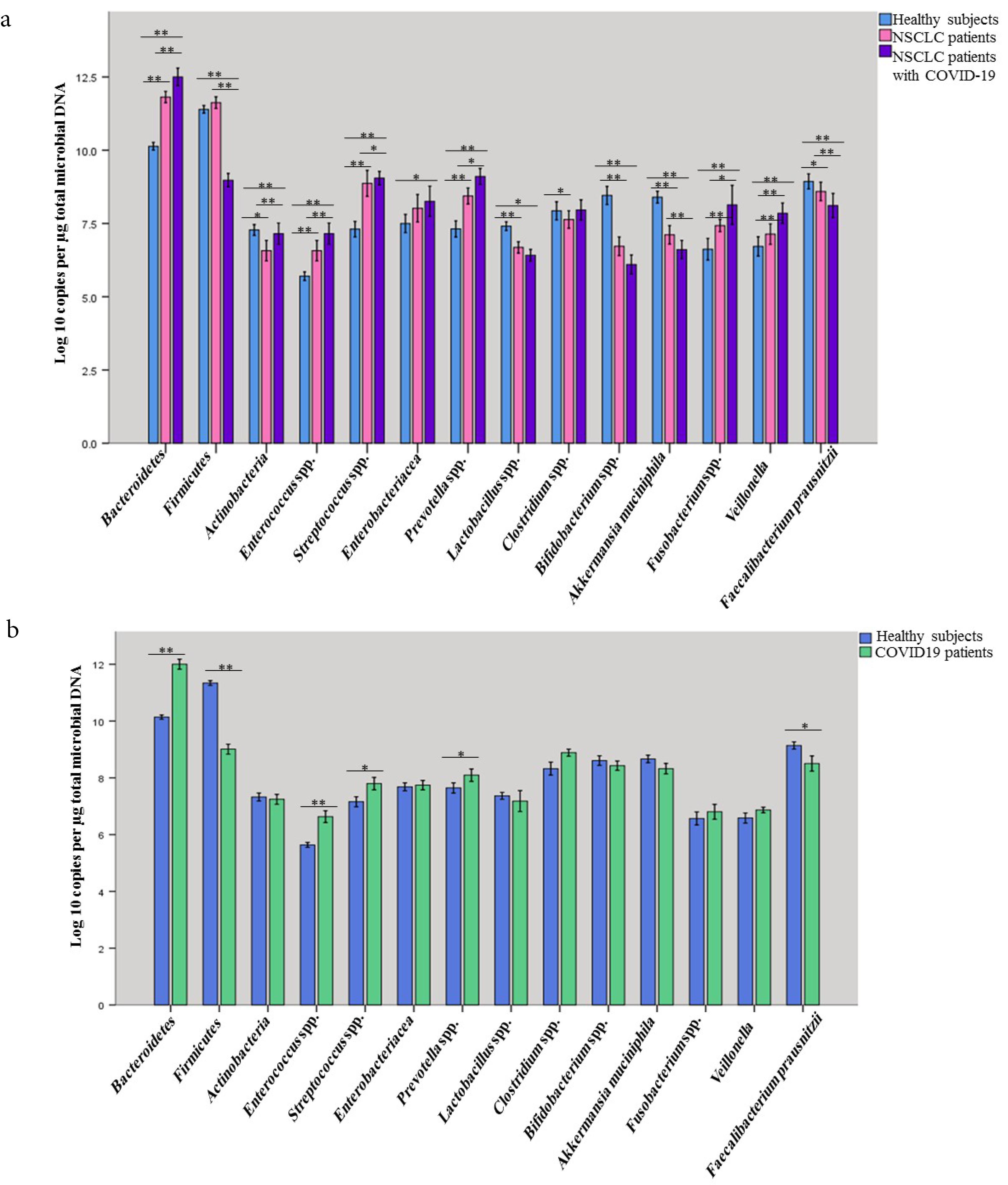
Figure 2.
(a) Comparing the presence of targeted members of gut microbiota of studied groups, including NSCLC (non-small cell lung cancer) patients, NSCLC patients with COVID-19 and healthy subjects. (b) Comparing the presence of targeted members of gut microbiota of studied groups, including COVID-19 patients and healthy subjects. Data of qPCR are presented as mean log10 copies/µg of total microbial DNA. In these diagrams, the error bar indicates mean ± SD, 95% CI, **P < 0.01, *P < 0.05
.
(a) Comparing the presence of targeted members of gut microbiota of studied groups, including NSCLC (non-small cell lung cancer) patients, NSCLC patients with COVID-19 and healthy subjects. (b) Comparing the presence of targeted members of gut microbiota of studied groups, including COVID-19 patients and healthy subjects. Data of qPCR are presented as mean log10 copies/µg of total microbial DNA. In these diagrams, the error bar indicates mean ± SD, 95% CI, **P < 0.01, *P < 0.05
Table 3.
Comparison of Population Numbers of Selected Microbial Groups in the Lung of Healthy Control, NSCLC Patients, and NSCLC Patients with COVID-19
|
Target Bacterial
|
Healthy Control
(n=40)
|
NSCLC Patients
(n=20)
|
NSCLC Patients with COVID-19 (n=17)
|
P
Value
|
|
A
|
B
|
C
|
| Bacteroidetes |
3140 ± 768.54 |
2235 ± 534.3 |
2470.5 ± 908.8 |
< 0.001 |
0.009 |
< 0.001 |
| Firmicutes |
4540.5 ± 1001.74 |
3655 ± 820.4 |
3900 ± 861.68 |
0.003 |
0.060 |
< 0.001 |
| Actinobacteria |
130 ± 37.96 |
112 ± 33.49 |
141.93 ± 29.74 |
0.197 |
0.295 |
< 0.001 |
|
Prevotella spp. |
2.12 ± 0.143 |
2.16 ± 0.166 |
2.26 ± 0.232 |
< 0.001 |
0. 261 |
0.026 |
|
Veillonella
|
114.2 ± 43.37 |
154 ± 36.03 |
133.11 ± 41.5 |
0.061 |
0.382 |
< 0.001 |
|
Streptococcus spp. |
126 ± 34.71 |
141 ± 47.56 |
157 ± 49.01 |
0.003 |
< 0.001 |
0.002 |
A, NSCLC patients vs. Healthy control; B, NSCLC patients with COVID-19 vs. Healthy control; C, NSCLC patients with COVID-19 vs. NSCLC patients.
Relationships among between the groups were performed assessed using the ANOVA test in multiple comparisons. Values were presented as mean ± standard deviation (SD). P values < 0.05 was considered statistically significant.
Table 4.
Copies/mL of Colonized Bacteria in Studied Patients (NSCLC Patients with COVID-19 and COVID-19 Patients Without NSCLC)
|
Bacterial
|
Patient type
|
≥102
copies/mL
No. (%)
|
≥102
-<105
copies/mL
No. (%)
|
≥105
copies/mL
No. (%)
|
|
S. aureus
|
NSCLC patients with COVID-19 (n = 17) |
1 |
- |
- |
| Patients with COVID-19 (n = 40) |
- |
1 |
1 |
|
S. pneumonia
|
NSCLC patients with COVID-19 (n = 17) |
- |
- |
- |
| Patients with COVID-19 (n = 40) |
- |
1 |
- |
|
M. catarrhalis
|
NSCLC patients with COVID-19 (n = 17) |
- |
1 |
- |
| Patients with COVID-19 (n = 40) |
- |
- |
- |
|
H. influenza
|
NSCLC patients with COVID-19 (N = 17) |
- |
- |
1 |
| Patients with COVID-19 (N = 40) |
- |
- |
- |
|
P. aeruginosa
|
NSCLC patients with COVID-19 (N = 17) |
2 |
- |
2 |
| Patients with COVID-19 (n = 40) |
1 |
2 |
- |
Data are presented as Log copies/mL. Absent (Log copies /mL < 102), Present (Log CFU/mL 102-105).
Quantitation of Fecal Microbiota by qPCR
Quantities of Bacteroidetes, Firmicutes, Actinobacteriaphyla, Enterococcus spp., Streptococcus spp., Enterobacteriaceae, Prevotella spp., Lactobacillus spp., Clostridium spp., Bifidobacterium spp., F. prausnitzii, Fusobacterium spp., A. muciniphila, andVeillonella were detected in all fecal specimens; however, the numbers of Bacteroidetes, Firmicutes, and Actinobacteriaphyla were significantly (P < 0.001, Table 5, Figure 3) higher among NSCLC patients with COVID-19 compared with COVID-19 patients and healthy subjects. NSCLC patients with COVID-19 had lower levels of Lactobacillus spp., A. muciniphila, and Bifidobacterium spp. compared to COVID-19 patients and healthy control group (P < 0.001). Our results confirmed that COVID-19 patients showed a significant decline in Firmicutes (P = 0.00), Prevotella spp., (P = 0.015), F. prausnitzii (P = 0.013) while increased quantities of Bacteroidetes, (P < 0.001), and Enterococcus spp., (P < 0.001), Streptococcus spp., (P = 0.021) were observed in comparison with healthy subjects (Figure 3 and Table S7). The counts of Streptococcus spp., in the lung and gut, had been notably (P < 0.05) higher in the NSCLC patients suffering from COVID-19 (8.49 ± 0.70 log CFU/g wet feces) than healthy controls (7.15 ± 0.55 log CFU/g wet feces) (Table 5, Supplementary Tables S6 and S7). Of note, Prevotella spp. was also enriched in the gut and respiratory tracts of COVID-19 patients with or without lung cancer (P < 0.05). The differences in intestinal bacterial of COVID-19 patients are illustrated in Figures S1 and S2.
Table 5.
Comparison of Population Numbers of Selected Microbial Groups in Healthy Control, NSCLC Patients, and NSCLC Patients with COVID-19
|
Target bacterial
|
Healthy Control
(n=40)
|
NSCLC Patients
(n=20)
|
NSCLC Patients
with COVID-19
(n=17)
|
P
Value
|
|
A
|
B
|
C
|
| Bacteroidetes |
10.13 ± 0.23 |
11.7 ± 0.37 |
12.5 ± 0.59 |
< 0.001 |
< 0.001 |
< 0.001 |
| Firmicutes |
11.34 ± 0.46 |
10.89 ± 0.19 |
8.97 ± 0.45 |
0.068 |
0 < 0.001 |
< 0.001 |
| Actinobacteria |
7.32 ± 0.34 |
7.05 ± 0.28 |
5.68 ± 0.25 |
0.034 |
< 0.001 |
< 0.001 |
|
Enterococcus spp. |
5.63 ± 0.27 |
6.53 ± 0.64 |
7.15 ± 0.70 |
< 0.001 |
< 0.001 |
< 0.001 |
|
Streptococcus spp. |
7.15 ± 0.55 |
7.9 ± 1.00 |
8.49 ± 0.70 |
< 0.001 |
< 0.001 |
0.015 |
|
Enterobacteriaceae
|
7.68 ± 0.43 |
7.92 ± 0.88 |
8.25 ± 1.00 |
0.658 |
0.023 |
0.522 |
|
Prevotella spp. |
7.64 ± 0.61 |
8.47 ± 0.15 |
9.10 ± 0.53 |
< 0.001 |
< 0.001 |
0.021 |
|
Lactobacillus spp.* |
3.04E7 ± 1.91E7 |
6.49E6 ± 5.27E6 |
3.78E6 ± 9.81E5 |
0.002 |
0.051 |
0.981 |
|
Clostridium spp. |
8.23 ± 0.71 |
7.52 ± 0.16 |
7.96 ± 0.67 |
0.05 |
0.207 |
0.158 |
|
Bifidobacterium spp. |
8.60 ± 0.53 |
6.5 ± 0.74 |
6.09 ± 0.63 |
< 0.001 |
0.003 |
0.112 |
|
F. prausnitzii
|
9.13 ± 0.40 |
8.57 ± 0.29 |
8.11 ± 0.80 |
0.05 |
< 0.001 |
0.003 |
|
Fusobacterium spp. |
6.56 ± 0.71 |
7.44 ± 0.43 |
8.13 ± 1.29 |
< 0.001 |
< 0.001 |
0.045 |
|
A. muciniphila
|
8.66 ± 0.41 |
7.16 ± 0.58 |
6.60 ± 0.61 |
< 0.001 |
< 0.001 |
0.004 |
|
Veillonella
|
6.58 ± 0.55 |
7.15 ± 0.63 |
7.84 ± 0.86 |
0.003 |
< 0.001 |
0.002 |
A, NSCLC patients vs. Healthy control; B, NSCLC patients without COVID-19 vs. Healthy control; C, NSCLC patients with COVID-19 vs. NSCLC patients
*Bacteria with normal distribution.
Relationships between the groups were assessed using the ANOVA test in multiple comparisons. Values were presented as mean ± standard deviation (SD). P values < 0.05 was considered statistically significant.
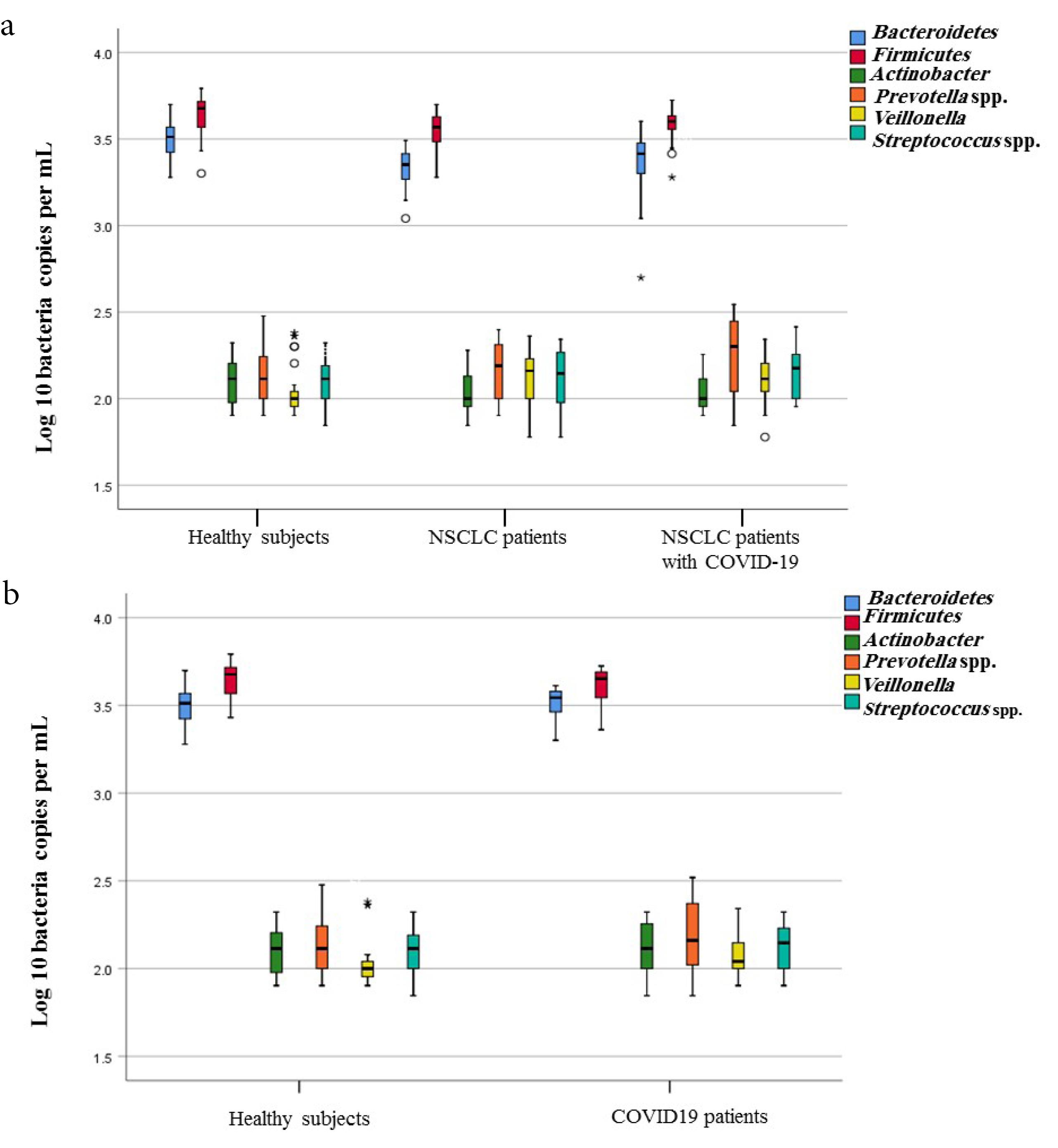
Figure 3.
Box-and-Whisker Plots of Bacterial Groups Quantified by qPCR. Bacterial groups quantified by SYBR Green qPCR and expressed as Log10 bacteria per mL nasopharyngeal in the lung microbiota of studied groups. (a) Comparing the presence of targeted members of lung microbiota of studied groups, including NSCLC (non-small cell lung cancer) patients, NSCLC patients with COVID-19 and healthy subjects. (b) Comparing the presence of targeted members of lung microbiota of studied groups, including COVID-19 patients and healthy subjects
.
Box-and-Whisker Plots of Bacterial Groups Quantified by qPCR. Bacterial groups quantified by SYBR Green qPCR and expressed as Log10 bacteria per mL nasopharyngeal in the lung microbiota of studied groups. (a) Comparing the presence of targeted members of lung microbiota of studied groups, including NSCLC (non-small cell lung cancer) patients, NSCLC patients with COVID-19 and healthy subjects. (b) Comparing the presence of targeted members of lung microbiota of studied groups, including COVID-19 patients and healthy subjects
Discussion
Characterization of the gut and lung microbiota in patients with NSCLC, as a common cancer, particularly when complicated by SARS-CoV-2 infection, is less known.3,13 However, learning about the systemic influence of the gut microbiota and its relation to the immune system made a huge leap in clarifying the influence of gut microbiota on chemotherapy and lung cancer treatment.3
We found that the rate of chemotherapy treatment before infection withSARS-CoV-2 in NSCLC patients was significantly higher than lung cancer patients. CT images in NSCLC patients with COVID-19 showed more ground-glass opacity than COVID-19 patients.
Our data in accordance with other studies revealed that in lung cancer patients, cumulative risk factors including chemotherapy and ICU admission correlated with severe outcomes in COVID-19 patients.13,14,17
In a healthy individual, the lung microbiota has a low population density but an important variety of interacting microbiota. Usually, there are species of the Firmicutes such as Streptococcus sp. and Veillonella sp., Bacteroidetes including Prevotella sp., and Proteobacteria phyla in the lung microbiota of healthy adult individuals.2,30 The lung microbiota is altered in NSCLC and the microbiota affects cancer development and progression.14
The abundance of Streptococcus spp., was higher in NSCLC patients with COVID-19; additionally, an increased quantity of Prevotella spp. was found in COVID-19 patients.Prevotella and Veillonella were most strongly associated with NSCLC, and Veillonella significantly intensified the progression of NSCLC in lung microbiota.17 Persistence of Veillonella sp. andPrevotella spp. in the airways, rise their metabolism of mucus and more colonization by bacterial pathogens causing pneumonia.31
Dysbiosis in the lung microbiota population such as a diminishment in Betaproteobacteria and an increment in Staphylococcus, Streptococcus, and Enterobacteriaceae correlates with the host antiviral immune response together with serum cytokine IL-6.12,14
Consequently, the lung microbiota predicts the severity of disease and clinical outcomes in these patients.14
The landscape of lung microbiota in COVID-19 patients showed that some pathogens, such as Klebsiella oxytoca causingpneumonia, colitis, and sepsis, are increased in COVID-19 patients and reduced in healthy individuals.12,32 According to the results, P. aeruginosa and S. aureus were the mostcommon pulmonary bacterial groups that colonized in the lung microbiota of COVID-19 patients.
Other studies using a high-throughput sequencing procedure in analyzing the oropharyngeal microbiota of patients with pneumonia have found a significant rise in Pseudomonas and Bacillus species after influenza viral infection; however, the number of Veillonella, Prevotella, and Neisseria were significantly decreased.33 In nasopharyngeal microbiota of COVID-19 patients, taxa such as Staphylococcus, that were detected in our study, were shown to increase in the nasopharynx microbiota of H3N2 influenza patients in comparison to control groups.34
As already detailed, there are differences between the population density and composition of the lung microbiota and that of the intestinal tract due to bidirectional movement of air and mucus14 Other independent studies showed that compared to healthy individuals, the diversity of the intestinal microbiota was significantly decreased in SARS COV-2 positive patients.35 Key taxa such as F. prausnitzii, Bifidobacteria sp., Actinobacteria, Ruminococcus sp., and Lachnospiraceae were depleted but the abundance of A. muciniphila, Bacteroides sp.,Veillonella, Clostridium sp., Enterococcus and Streptococcus was increased.1,36 The stools of these NSCLC patients were revealed to be enriched in phylum Firmicutes, Akkermansia, and Ruminococcus (F. prausnitzii) genera.3
Similar to another previous study, a reduction in bacterial frequency was revealed in stool samples of COVID-19 patients compared with the healthy control group. Our evidence of the intestinal microbiota indicated a significant decline in Firmicutes, Prevotella spp., and F. prausnitzii in COVID-19 patients compared to the control groups. The intestinal bacteria could transfer to other organs through the intestinal barrier, and the COVID-19 infection could promote the switch of intestinal bacteria.32
Faecalibacterium prausnitzii produces short chain fatty acids (SCFAs) which play an important role in neuro-immunoendocrine regulation and are key sources for intestinal epithelial cells to improve the gut barrier function37 Besides, SCFAs activate signaling cascade by cell surface G-protein coupled receptors that control immune functions and inflammatory side effects.1 F. prausnitzii, which produces butyrate and IL-10 in the gut leads to a negative relation with COVID-19 severity and protects the host from the infection.12,32
Consistent with the gut-lung axis concept, a cross-sectional study showed that the bacterial diversity of gut microbiota is significantly decreased in COVID-19 patients (with and without NSCLC). In our study, increased quantities of Streptococcus spp., and Prevotella spp. (P <0.05) were found in the gut-lung axis of NSCLC patients with COVID-19 (with and without NSCLC).
Prevotella genusis commensal and scarcely ever involved in acute infections. However, some strains are opportunistic pathogens in chronic diseases, mucosal inflammation, and SARS-CoV-2 with activation of immune signaling pathways.31 The bacterial population also suggests the robustness of the lung bacterial community in limiting SARS-CoV-2 increase or attachment.
An increased levels of bacteria belonging toFirmicutes, Actinobacteria, and Bacteroidetes phyla (P < 0.001), which produce SCFAs, particularly butyric acid.12 A lower plentitude of Lactobacillus spp., A. muciniphila, and Bifidobacterium spp.,was observed in NSCLC patients with COVID-19 when compared with control subjects. As stated previously, dysbiosis of the gut- lung axis including Bifidobacterium and Lactobacillus communities can increase the abundances of opportunistic pathogensin COVID-19 and systemic inflammatory response syndrome patients.37,38 Furthermore, patients with NSCLC or renal cell carcinoma responding to immunotherapy showed enhancement of A. muciniphila at diagnosis.39
Woodall et al reported increased amounts of Enterococcus in the gut microbiota of patients with COVID-19 which is similar to this survey.1 Many strains of Enterococcus species are pathobionts and have been shown in SARS-CoV-2 infection, gastric and colorectal cancers.35
A recent cohort study in COVID-19 patients revealed persistent microbial dysbiosis with previous viral clearance, regardless of antibiotics and anti-viral medications.40 Gut-colonizing bacteria such as Enterobacteriales in the respiratory tract have detrimental effects on the outcomes of respiratory diseases but increased gut-colonizing organisms were not discovered in this study.8 NSCLC patients who responded to neoadjuvant and chemotherapy agents harbored a higher diversity of gut microbiota at baseline with constant composition during therapy.41
Some limitations of the study are as follows: Obtaining NSCLC specimens from different groups of studied patients was time-consuming; therefore, the sample size of the study should be increased in future researches. Because of limited resources of NGS technologies in developing countries, we performed the qRT-PCR based on different targeted bacteria quantities. The gut microbiota plays a crucial function in host homeostasis and promises a new anti-lung cancer approach, but there is a need to further evaluate lung microbiota and its products, both in health and in disease.
The studied patients showed evidence of nasopharyngeal microbiota disorders connected with disease severity, and there was a significant relation between severity of COVID-19 and microbiota signatures early in the disease. There are significant relations between the duration of hospitalization, the type of oxygen support, and antibiotic treatment, which affect the microbiota of the upper respiratory tract. One of the weaknesses of the current study is that we could not measure confounding variables and adjust their effect in the model; therefore, subsequent studies should control the influence of these confounders.
Because of the ambiguous extent of the contribution of the human microbiota to COVID-19 and lung cancer, further microbiota studies from different countries, severe clinical conditions, and longitudinal information (e.g. post-hospitalization) are required.
Conclusion
Finally, we found that compared to control groups, COVID-19 patients with NSCLC showed a diminishment in gut bacteria diversity and increase in Lactobacillus spp., A. muciniphila, and Bifidobacterium spp. Unbiased analysis indicated an increment in Enterococcus spp., Streptococcus spp., and Prevotella spp. in COVID-19 patients with NSCLC. Distinctive microbiota strategies and confounder effects lead to high heterogeneity across studies,which showed lower plenitude in COVID-19 patients with NSCLC than control subjects.
Supplementary Files
Supplementary file 1 contains Tables S1-S7 and Figures S1 and S2.
(pdf)
Acknowledgements
The staff of Omid Hospital is highly appreciated for their great aid in gathering the required samples. The authors would like to appreciate Dr. Peyman Adibi, and Dr. Bahram Bagherpour for their cooperation in our research.
Competing Interests
The authors have no conflicts of interest to declare regarding this paper.
Ethical Approval
The research project was approved by the human research ethics committee of Isfahan University of Medical Sciences (Permit Number 52056) and followed the guidelines established in the Declaration of Helsinki (2000). All participants in this study supplied their informed consent in writing.
Funding
The current study was supported by a grant (No, 52056) from Isfahan University of Medical Sciences, Isfahan, Iran.
References
- Woodall CA, McGeoch LJ, Hay AD, Hammond A. Respiratory tract infections and gut microbiome modifications: a systematic review. PLoS One 2022; 17(1):e0262057. doi: 10.1371/journal.pone.0262057 [Crossref] [ Google Scholar]
- Yuksel N, Gelmez B, Yildiz-Pekoz A. Lung microbiota: its relationship to respiratory system diseases and approaches for lung-targeted probiotic bacteria delivery. Mol Pharm 2023; 20(7):3320-37. doi: 10.1021/acs.molpharmaceut.3c00323 [Crossref] [ Google Scholar]
- Bingula R, Filaire M, Radosevic-Robin N, Berthon JY, Bernalier-Donadille A, Vasson MP. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma: study protocol for case-control observational trial. Medicine (Baltimore) 2018; 97(50):e13676. doi: 10.1097/md.0000000000013676 [Crossref] [ Google Scholar]
- Al Kassaa I, El Omari S, Abbas N, Papon N, Drider D, Kassem Kassem, II II. High association of COVID-19 severity with poor gut health score in Lebanese patients. PLoS One 2021; 16(10):e0258913. doi: 10.1371/journal.pone.0258913 [Crossref] [ Google Scholar]
- Dhar D, Mohanty A. Gut microbiota and COVID-19- possible link and implications. Virus Res 2020; 285:198018. doi: 10.1016/j.virusres.2020.198018 [Crossref] [ Google Scholar]
- He ZJ, Liang YX, Cai LY. Advances in the interaction between intestinal microbiota and COVID-19. Explor Res Hypothesis Med 2021; 6(1):1-8. doi: 10.14218/erhm.2020.00055 [Crossref] [ Google Scholar]
- Zhang D, Li S, Wang N, Tan HY, Zhang Z, Feng Y. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol 2020; 11:301. doi: 10.3389/fmicb.2020.00301 [Crossref] [ Google Scholar]
- Bao L, Zhang C, Dong J, Zhao L, Li Y, Sun J. Oral microbiome and SARS-CoV-2: beware of lung co-infection. Front Microbiol 2020; 11:1840. doi: 10.3389/fmicb.2020.01840 [Crossref] [ Google Scholar]
- McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur J Immunol 2018; 48(1):39-49. doi: 10.1002/eji.201646721 [Crossref] [ Google Scholar]
- Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem?. Front Microbiol 2016; 7:455. doi: 10.3389/fmicb.2016.00455 [Crossref] [ Google Scholar]
- Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020; 5(5):434-5. doi: 10.1016/s2468-1253(20)30083-2 [Crossref] [ Google Scholar]
- Yamamoto S, Saito M, Tamura A, Prawisuda D, Mizutani T, Yotsuyanagi H. The human microbiome and COVID-19: a systematic review. PLoS One 2021; 16(6):e0253293. doi: 10.1371/journal.pone.0253293 [Crossref] [ Google Scholar]
- Nie L, Dai K, Wu J, Zhou X, Hu J, Zhang C. Clinical characteristics and risk factors for in-hospital mortality of lung cancer patients with COVID-19: a multicenter, retrospective, cohort study. Thorac Cancer 2021; 12(1):57-65. doi: 10.1111/1759-7714.13710 [Crossref] [ Google Scholar]
- Khatiwada S, Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Hum Microb J 2020; 17:100073. doi: 10.1016/j.humic.2020.100073 [Crossref] [ Google Scholar]
- Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis 2020; 71(10):2669-78. doi: 10.1093/cid/ciaa709 [Crossref] [ Google Scholar]
- Fan J, Li X, Gao Y, Zhou J, Wang S, Huang B. The lung tissue microbiota features of 20 deceased patients with COVID-19. J Infect 2020; 81(3):e64-7. doi: 10.1016/j.jinf.2020.06.047 [Crossref] [ Google Scholar]
- Zeng W, Zhao C, Yu M, Chen H, Pan Y, Wang Y. Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered 2022; 13(3):6665-77. doi: 10.1080/21655979.2022.2045843 [Crossref] [ Google Scholar]
- Rogado J, Pangua C, Serrano-Montero G, Obispo B, Marino AM, Pérez-Pérez M. COVID-19 and lung cancer: a greater fatality rate?. Lung Cancer 2020; 146:19-22. doi: 10.1016/j.lungcan.2020.05.034 [Crossref] [ Google Scholar]
- Mostafa HH, Fissel JA, Fanelli B, Bergman Y, Gniazdowski V, Dadlani M. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio 2020; 11(6):e01969-20. doi: 10.1128/mBio.01969-20 [Crossref] [ Google Scholar]
- Yang K, Sheng Y, Huang C, Jin Y, Xiong N, Jiang K. Clinical characteristics, outcomes, and risk factors for mortality in patients with cancer and COVID-19 in Hubei, China: a multicentre, retrospective, cohort study. Lancet Oncol 2020; 21(7):904-13. doi: 10.1016/s1470-2045(20)30310-7 [Crossref] [ Google Scholar]
- Nasri E, Shoaei P, Vakili B, Mirhendi H, Sadeghi S, Hajiahmadi S. Fatal invasive pulmonary aspergillosis in COVID-19 patient with acute myeloid leukemia in Iran. Mycopathologia 2020; 185(6):1077-84. doi: 10.1007/s11046-020-00493-2 [Crossref] [ Google Scholar]
- Laroumagne S, Salinas-Pineda A, Hermant C, Murris M, Gourraud PA, Do C. [Incidence and characteristics of bronchial colonisation in patient with lung cancer: a retrospective study of 388 cases]. Rev Mal Respir 2011; 28(3):328-35. doi: 10.1016/j.rmr.2010.05.020.[French] [Crossref] [ Google Scholar]
- Shoaei P, Shojaei H, Siadat SD, Moshiri A, Vakili B, Yadegari S. Gut microbiota in burned patients with Clostridioides difficile infection. Burns 2022; 48(5):1120-9. doi: 10.1016/j.burns.2021.11.023 [Crossref] [ Google Scholar]
- Wu D, Hou C, Li Y, Zhao Z, Liu J, Lu X. Analysis of the bacterial community in chronic obstructive pulmonary disease sputum samples by denaturing gradient gel electrophoresis and real-time PCR. BMC Pulm Med 2014; 14:179. doi: 10.1186/1471-2466-14-179 [Crossref] [ Google Scholar]
- Koliada A, Syzenko G, Moseiko V, Budovska L, Puchkov K, Perederiy V. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 2017; 17(1):120. doi: 10.1186/s12866-017-1027-1 [Crossref] [ Google Scholar]
- Yang YW, Chen MK, Yang BY, Huang XJ, Zhang XR, He LQ. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol 2015; 81(19):6749-56. doi: 10.1128/aem.01906-15 [Crossref] [ Google Scholar]
- Silva-Junior WP, Martins AS, Xavier PC, Appel KL, Oliveira Junior SA, Palhares DB. Etiological profile of early neonatal bacterial sepsis by multiplex qPCR. J Infect Dev Ctries 2016; 10(12):1318-24. doi: 10.3855/jidc.7474 [Crossref] [ Google Scholar]
- Curran T, Coyle PV, McManus TE, Kidney J, Coulter WA. Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol 2007; 50(1):112-8. doi: 10.1111/j.1574-695X.2007.00241.x [Crossref] [ Google Scholar]
- Ohkusu K, Nash KA, Inderlied CB. Molecular characterisation of Haemophilus influenzae type a and untypeable strains isolated simultaneously from cerebrospinal fluid and blood: novel use of quantitative real-time PCR based on the cap copy number to determine virulence. Clin Microbiol Infect 2005; 11(8):637-43. doi: 10.1111/j.1469-0691.2005.01203.x [Crossref] [ Google Scholar]
- Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol 2016; 78:481-504. doi: 10.1146/annurev-physiol-021115-105238 [Crossref] [ Google Scholar]
- Ventero MP, Cuadrat RR, Vidal I, Andrade BG, Molina-Pardines C, Haro-Moreno JM. Nasopharyngeal microbial communities of patients infected with SARS-CoV-2 that developed COVID-19. Front Microbiol 2021; 12:637430. doi: 10.3389/fmicb.2021.637430 [Crossref] [ Google Scholar]
- Han Y, Jia Z, Shi J, Wang W, He K. The active lung microbiota landscape of COVID-19 patients. medRxiv [Preprint]. August 23, 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.08.20.20144014v1.
- Zemanick ET, Wagner BD, Sagel SD, Stevens MJ, Accurso FJ, Harris JK. Reliability of quantitative real-time PCR for bacterial detection in cystic fibrosis airway specimens. PLoS One 2010; 5(11):e15101. doi: 10.1371/journal.pone.0015101 [Crossref] [ Google Scholar]
- De Maio F, Posteraro B, Ponziani FR, Cattani P, Gasbarrini A, Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biol Proced Online 2020; 22:18. doi: 10.1186/s12575-020-00131-7 [Crossref] [ Google Scholar]
- Ren Z, Wang H, Cui G, Lu H, Wang L, Luo H. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021; 70(7):1253-65. doi: 10.1136/gutjnl-2020-323826 [Crossref] [ Google Scholar]
- Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne) 2020; 11:25. doi: 10.3389/fendo.2020.00025 [Crossref] [ Google Scholar]
- Haldar S, Jadhav SR, Gulati V, Beale DJ, Balkrishna A, Varshney A. Unravelling the gut-lung axis: insights into microbiome interactions and Traditional Indian Medicine’s perspective on optimal health. FEMS Microbiol Ecol 2023; 99(10):fiad103. doi: 10.1093/femsec/fiad103 [Crossref] [ Google Scholar]
- Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology 2020;159(4):1302-10.e5. 10.1053/j.gastro.2020.06.048.
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359(6371):91-7. doi: 10.1126/science.aan3706 [Crossref] [ Google Scholar]
- Newsome RC, Gauthier J, Hernandez MC, Abraham GE, Robinson TO, Williams HB. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes 2021; 13(1):1-15. doi: 10.1080/19490976.2021.1926840 [Crossref] [ Google Scholar]
- Nagasaka M, Sexton R, Alhasan R, Rahman S, Azmi AS, Sukari A. Gut microbiome and response to checkpoint inhibitors in non-small cell lung cancer-a review. Crit Rev Oncol Hematol 2020; 145:102841. doi: 10.1016/j.critrevonc.2019.102841 [Crossref] [ Google Scholar]