Arch Iran Med. 25(12):798-806.
doi: 10.34172/aim.2022.125
Original Article
Association Between Human Leukocyte Antigens and Graft-Versus-Host Disease Occurrence in Allogeneic Hematopoietic Stem Cell Transplantation – A 10-Year Experience on Iranian Patients
Mohsen Hamidpour Conceptualization, Methodology, 1 
Elham Roshandel Investigation, Writing – review & editing, 1 
Haniyeh Ghaffari Nazari Investigation, Writing – original draft, 1
Ghazaleh Sankanian Data curation, Investigation, 1
Hossein Bonakchi Data curation, Formal analysis, 1
Maryam Salimi Investigation, 1
Sina Salari Conceptualization, Supervision, 1, * 
Author information:
1Hematopoietic Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Abstract
Background:
Human leukocyte antigen (HLA) molecules mediate critical roles in determining responsiveness or non-responsiveness of the immune system, especially in transplantation. Some studies have shown a possible association between certain HLA alleles and some allogeneic hematopoietic stem cell transplantation (allo-HSCT) outcomes such as acute/chronic graft-versus-host disease (aGVHD/cGVHD) and overall survival (OS). In the current study, we investigated any possible association of HLA subclasses and acute/chronic GVHD occurrence as well as OS in patients receiving HLA-matched sibling allo-HSCT.
Methods:
We retrospectively evaluated the association of various HLA alleles with the incidence of aGVHD, cGVHD, and OS of 162 patients who received allo-HSCT from HLA-matched sibling between 2009-2018 at Taleghani hospital in Tehran.
Results:
We found that the incidence of aGVHD grades II-IV was higher among patients who had HLA-B*07 (P=0.031) and HLA-DRB1*07 (P=0.052). The presence of HLA-A*01 was associated with 4.5-fold greater odds of incidence in the extensive-type of cGVHD (P=0.009). Furthermore, HLA-A*03 (P=0.089), HLA-B*13(P=0.013), HLA-B*40 (P=0.042), HLA-DRB1*02 (P=0.074), and HLA-DRB1*04 (P=0.039) were associated with a lower rate of OS.
Conclusion:
This study suggests that certain HLA alleles might influence the incidence and severity of acute or chronic GVHD in the context of HLA-matched sibling allo-HSCT. In addition, some specific HLA alleles help predict OS in allo-HSCT recipients. These results might be helpful in estimating the incidence of aGVHD, cGVHD, and OS as well as designing personalized therapy.
Keywords: Allogeneic hematopoietic stem cell transplantation, Graft-versus-host disease, Human leukocyte antigen, Survival
Copyright and License Information
© 2022 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article as: Hamidpour M, Roshandel E, Ghaffari Nazari H, Sankanian G, Bonakchi H, Salimi M, et al. Association between human leukocyte antigens and graft-versus-host disease occurrence in allogeneic hematopoietic stem cell transplantation – a 10-year experience on Iranian patients. Arch Iran Med. 2022;25(12):798-806. doi: 10.34172/aim.2022.125
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a promising therapy for many hematological malignancies and disorders.1 Despite the advances in pre- and post-transplant supportive care, allo-HSCT remains associated with substantial transplant-related mortality. Graft-versus-host disease (GVHD) represents the highest frequency in mortality following allo-HCT.2 Host tissue damage induced by conditioning regimens releases inflammatory cytokines and activates various subsets of immune cells, including antigen presenting cells, T cells, and natural killer cells, which are involved in the pathophysiology of GVHD.3 Donor immune cells that exist in grafts from different stem cell sources such as bone marrow, mobilized peripheral blood, and cord blood can induce GVHD reactions after transplantation. Moreover, allo-reactive T-cells are known as a dominant subset of immune cells to moderate GVHD. CD4 + T and CD8 + T cells are two main allo-reactive T cell subtypes which recognize exogenous and endogenous molecules presented on the major histocompatibility complex (MHC) class II and MHC class I molecules, respectively.3 MHC, also called human leukocyte antigen (HLA), is the most polymorphic protein in the body with many various subclasses in each population.4 Regarding their frequent expression in all nucleated cells and high immunogenicity, HLAs are the most important antigens in transplantation. In case of HLA miss-match transplantations, they can serve as a foreign antigen provoking immune reactions causing graft-failure or GVHD.5 Therefore, matched HLA antigens at A, B, C, DRB1, and DQB1 locus between recipient and donor as well as prescribing immunosuppressive regimens have been considered to limit GVHD occurrence.6 In spite of this strategy, GVHD occurs in approximately 30% of the patients who undergo allo-HSCT from HLA-matched related donors. This rate is roughly 70% for patients with HLA-matched unrelated donors.7 With regards to GVHD developing even in the HLA-identical sibling allo-HSCTs, some studies evaluated the possible association between the presence or absence of specific HLA antigens and GVHD incidence in HLA-matched related and unrelated donors. More frequent HLA alleles which either increase or decrease the incidence of GVHD in HLA-identical sibling transplantations were reported in various populations.8,9 However, some studies did not find any change in the risk of GVHD through HLA antigens after bone marrow transplantation from HLA-identical sibling.10 In the current study, we investigated any possible association of HLA subclasses and acute/chronic GVHD occurrence as well as OS in patients receiving HLA-matched sibling allo-HSCT.
Materials and Methods
Study Design and Data Collection
We revised the clinical data of patients who underwent allo-HSCT between 2009 and 2018 at Taleghani hospital, Tehran, Iran. Patients were excluded ifthey underwent a second HSCT or related-/unrelated-HLA-mismatched transplantation. One hundred and sixty-two patients were found who received allo-HSCT from HLA-matched siblings and were aged between 15–62 years. HLA alleles among GVHD and non-GVHD patients were assessed to find frequent alleles present in patients with GVHD. All relevant demographic and clinical data were obtained from clinical records.
HSCT Procedure and GVHD Diagnosis
Transplant procedures, including peripheral blood hematopoietic stem cell collection, conditioning regimen, GVHD prophylaxis, and supportive care (anti-microbial prophylaxis and transfusion) were performed for all patients as previously published.11-13 All recipients received G-CSF-induced peripheral blood stem cells from full HLA-matched sibling donors.
Conditioning regimens consisted of intravenous (IV) busulfan (0.8 mg/kg every 6 hours for 4 days) followed by either cyclophosphamide (60 mg/kg/d for 2 days) or fludarabine (30 mg/m2 of body surface area once a day for 5 days). The combination of fludarabine (30 mg/m2 for 5 days, IV), 1-(2-chloroethyl)-3-cyclohexyl-1-nitrosourea (CCNU) (100 mg/m2 for 2 days, oral) and melphalan (40 mg/m2 for one day, IV) was prescribed in patients with Hodgkin’s disease (HD) and non-Hodgkin’s lymphoma (NHL). In patients with aplastic anemia (AA) and Fanconi anemia (FA), the administrated conditioning regimen was cyclophosphamide (as mentioned above) and 1.5 mg/kg of anti-thymocyte globulin (ATG) for three days.
GVHD prophylaxis containing cyclosporine A (CsA), and methotrexate (MTX) was given daily starting two days before transplantation. All patients received 3 mg/kg/d, IV of CsA from day -2 until + 5 (the day of allo-HSCT is considered day 0) followed by 12.5 mg/kg/d, P.O. until day + 180. Then, 10 mg/kg MTX, IV was added to the regimen from day + 1, with a dosage change to 6 mg/kg on days + 3, + 6 and + 11. In some patients, a dose of 1.5 mg/kg ATG, IV for 3 days (-3 until -1) was included in the combination.
GVHD was diagnosed and graded according to the National Institutes of Health (NIH) and modified Glucksberg criteria2,14 and treated with adjusted CsA dose and methylprednisolone as the first line of treatment. Infection prophylaxis was comprised of acyclovir (antiviral), ciprofloxacin (antibacterial), and fluconazole (antifungal). Grade 0 and I of aGVHD was considered as no GVHD and mild GVHD with slight transient symptoms, respectively. Grade II-IV of aGVHD was considered as moderate to severe aGVHD. Extensive cGVHD was defined as generalized and/or localized skin involvement skin or liver dysfunction plus involvement of other target organs. The hemoglobin threshold was considered 7g/dL for RBC transfusion. The PLT cut-off for platelet transfusion was 20 × 109/L.
HLA Typing
The DNA-based HLA typing technique determined HLA allele types in both patients and donors. Genomic DNA was extracted from blood samples collected with EDTA by the sailing out method (Yekta Tajhiz, Iran).15 The quantification of DNAs was measured using a 260 nm spectrophotometer, and then DNA samples were stored at -70oC. Specific primer-polymerase chain reaction (SSP-PCR) (Olerup commercial kit, Sweden) was applied to characterize HLA-A, B, and DRB1in two digits. The SSP steps are as follows;
simple
-
1- Initial denaturation for 2 minutes at 94°C
-
2- Denaturation for 10 seconds at 94°C
-
3- Annealing and extension for 1 minute at 65°C
Repeat steps 2 to 3, 10 times
simple
-
4- Denaturation for 10 seconds at 94°C
-
5- Annealing for 50 seconds at 61°C
-
6- Extension for 30 seconds at 72°C
Repeat steps 4 to 6, 20 times
simple
-
7- End-stage was held at RT if the process lasted less than 8 hours or at 4°C if longer than 8 hours. Final volume of PCR reactions was 10 µL
According to the manufacturer, agarose gel (2%) electrophoresis was used to resolve PCR products based on their molecular weight, visualized by staining with a safe stain under UV light, and photographed. SSP-PCR results were interpreted based on comparison with internal control relative sizes by data sheets of kit manufacture and software (SCORE version 6, Sweden).
Statistical Analyses
Analysis was conducted to determine the effect of risk factors and HLA-alleles on the acute and chronic GVHD using a logistic regression model. Overall survival (OS) was defined as the time from HSCT to death for any reason. Moreover, an analysis was performed to identify the effects of HLA-alleles on time to event using the Cox proportional hazard model. Plotting the score process and Kolmogorov-type supremum test were applied to execute the proportional hazards assumption. The significance level was 0.05.
In the univariable analysis for Cox and logistic regression, the significance level was set at 25%. In all of the multivariable analyses, a backward method with a significance level of 10% was utilized for selecting the highest predictive value features. The analysis was performed using the SAS program (version 9.4; SAS Institute Inc., Cary, NC, USA).
Results
Patients and Donors
The patients’ and donors’ clinical characteristics are shown in Table 1. The median age of the patients was 32.5 years (range: 1-62) of whom 83 (51.2%) and 78 (48.1%) were males and females, respectively. The median age of the donors was 32 years (range: 2-73) including 96 (59.35%) males and 64 (39.5%) females, respectively. Nine (5.6%), 10 (6.2%), 81 (50%), 42 (25.9%), 5 (3.1%) and 6 (3.7%) patients were diagnosed with NHL, HD, AML, ALL, AA and unclassified, respectively.
Table 1.
Clinical and Demographic Characteristics of Patients (N = 162).
|
Characteristics of Patients
|
|
| Recipient age (y), Median (Rang), Missing |
32.5 (15–62), 4 (2.5%) |
| Donor Age (y), Missing |
32 (18–73), 46 (28.4%) |
| Male:Female, Missing |
|
| Recipient |
83 (51.2%):78 (48.1%), 1 (0.6%) |
| Donor |
96 (59.3%):64 (39.5%), 2 (1.2%) |
| Gender match, Donor → Recipient |
|
| Male → Male |
45 (27.8) |
| Male → Female |
49 (30.2) |
| Female → Female |
26 (16) |
| Female → Male |
39 (24.1) |
| Missing |
3 (1.9) |
| Disease |
|
| NHL |
9 (5.6%) |
| HD |
10 (6.2%) |
| AML |
81 (50%) |
| ALL |
42 (25.9%) |
| Aplastic/Fanconi anemia |
5 (3.1%) |
| Unclassified |
6 (3.7%) |
| Missing |
9 (5.6%) |
| Recipient CMV IgG |
|
| Positive |
72 (44.4%) |
| Negative |
11 (6.8%) |
| Missing |
79 (48.8%) |
| Conditioning regimen |
|
| Bu/Cy |
85 (52.5%) |
| Bu/Flu |
37 (22.8%) |
| Bu/Flu/ATG |
15 (9.3%) |
| Flu/CCNU/Melphalan |
19 (11.7%) |
| Cy/ATG |
5 (3.1%) |
| Missing |
1 (0.6%) |
| Prophylaxis regimen |
|
| CSA + MTX |
102 (63%) |
| CSA + MTX + ATG |
16 (9.9%) |
| Missing |
44 (27.2%) |
| Compatibility blood group |
|
| Compatible |
85 (52.5%) |
| Incompatible |
71 (43.8%) |
| Missing |
6 (3.7%) |
| Delivery history (n = 64*) |
|
| Yes |
23 (35.9%) |
| No |
39 (60.9%) |
| Missing |
2 (3.1%) |
| Acute GVHD |
|
| 0-I |
123 (76%) |
| II-IV |
30 (18.5%) |
| Missing |
9 (5.5%) |
| Chronic GVHD |
|
| No cGVHD |
118 (72.8%) |
| Limited cGVHD |
4 (2.5%) |
| Extensive cGVHD |
5 (3.1%) |
| Missing |
35 (21.6%) |
*Number of Female Donors.
The Effect of HLA Alleles and Risk Factors on Acute and Chronic GVHD Incidence
In this center, the incidence of acute and chronic GVHD were 30 out of 153 (19.6%) and 9 out of 127 (7.1%) available records. The frequency distribution of HLA variants in patients is illustrated in Figure 1. In HLA-A, the highest frequency pertained to A*02 (22.2%) and the lowest frequencies pertained to A*34 and A*66 with 0.4%. In HLA-B, the highest frequency pertained to B*35 (18%) and the lowest frequencies pertained to B*14, B*32 with 0% and B*06, B*37, B*48, B*53 with 0.4%.
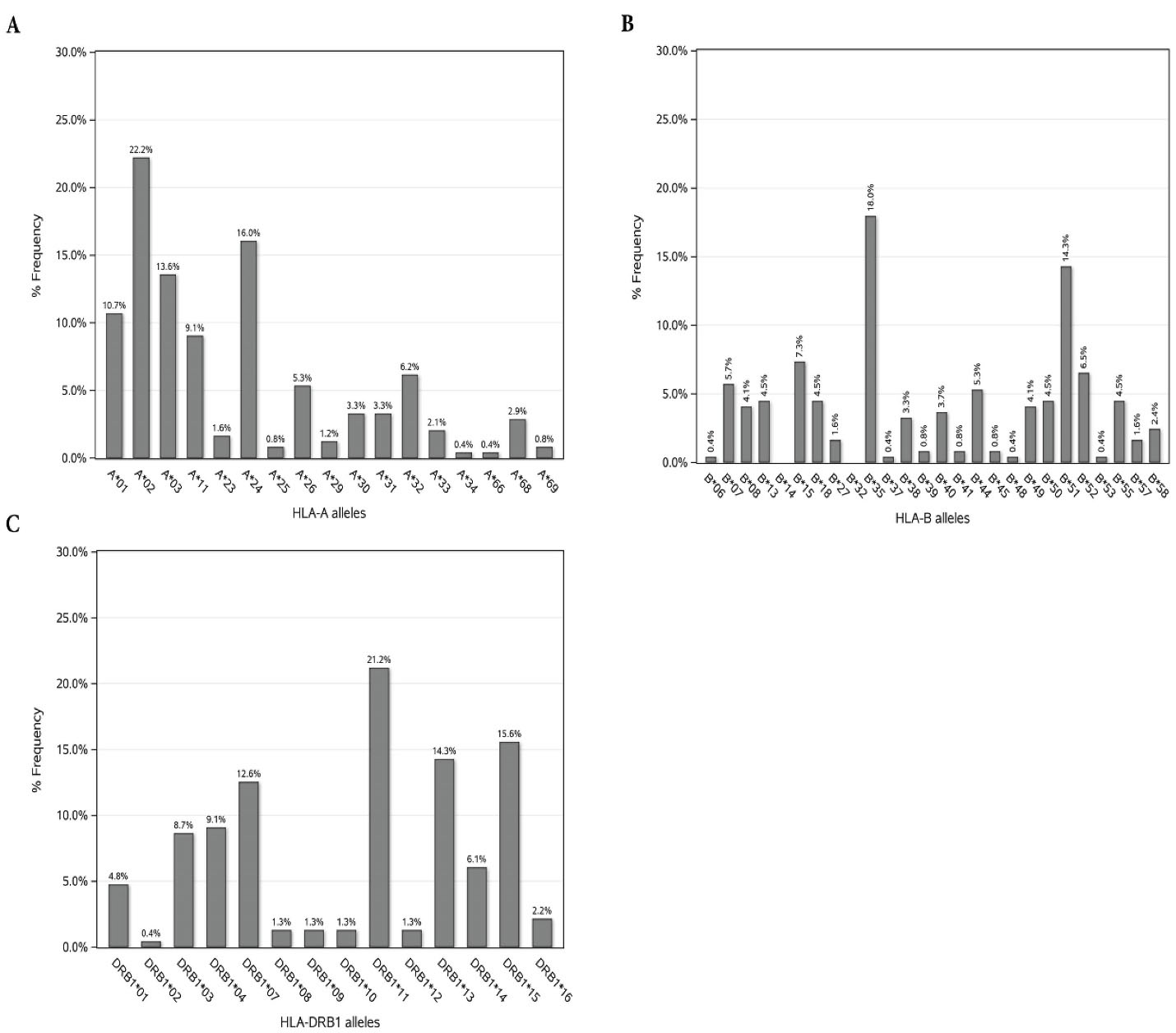
Figure 1.
Frequency of HLA -A, -B, and -DRB1 Alleles in HSCT Patients with HLA-Identical Sibling Donors
.
Frequency of HLA -A, -B, and -DRB1 Alleles in HSCT Patients with HLA-Identical Sibling Donors
In HLA-DRB1, the highest frequency pertained to DRB1-11 (21.2%) and the lowest frequency pertained to DRB1-02 (0.4%). The results for the effects of HLA alleles and the risk factors on acute and chronic GVHD along with their cross tabulation with occurrence acute and chronic GVHD are shown in Tables 2 and 3, respectively. As stated in Table 2, based on the univariate model, 9 HLA alleles were significantly associated with the incidence of grades II-IV acute GVHD. Also, CMV serostatus, gender and gender disparity between donor and recipient were significantly related to the incidence of grades II-IV acute GVHD. HLA-A*29, HLA-B*07, HLA-B*18, HLA-B*57, HLA-DRB1*07 and HLA-DRB1*13 were associated with the increase in incidence of grades II-IV acute GVHD; among which, A*29 and B*57 alleles caused a greater increase in the odds of acute GVHD incidence compared to other alleles (OR: 2.76; 75% CI: 1.35-5.66; P = 0.098). HLA-B*51, HLA-B*52 and HLA-DRB1*15 had a significant decreasing impact on the incidence of grades II-IV acute GVHD. The CMV positive patients had 7% greater odds of acute GVHD incidence compared to CMV negative patients (OR: 1.07; 75% CI: 1.13-2.56; P = 0.131). The female patients, in comparison with males, had 30% greater odds of acute GVHD incidence (OR: 1.30; 75% CI: 1.03-1.65; P value = 0.194). Transplantations with a male donor and a female recipient had 71% greater odds of grades II-IV acute GVHD incidence compared to male-to-male allo-HSCTs [OR: 1.71; 75%CI:(1.18-2.48); P value = 0.089]. According to the multiple model, HLA-B*07 and HLA-DRB1*07 were significantly associated with the incidence of grades II-IV acute GVHD in which by holding the effect of other variables constant, the presence of HLA-B*07 was associated with 3.84-fold increased odds of incidence (AOR: 3.84; 90% CI: 1.37-10.72; P = 0.031). Additionally, HLA-DRB1*07 was associated with 2.11-fold increased odds of incidence (AOR: 2.11; 90% CI: 1.12-3.95; P = 0.052). As detailed in Table 3, based on the univariate model, seven HLA alleles were significantly associated with the incidence of the extensive-type of chronic GVHD. The presence of B*40, B*41, B*57 alleles increased the odds of extensive-type chronic GVHD incidence more than other alleles (OR: 4.69; 75% CI: 1.97-11.13; P = 0.029]. Based on the multiple model, HLA-A*01 was significantly associated with the incidence of chronic GVHD and the patients with A*01 allele had 4.5-fold greater odds of extensive-type incidence of chronic GVHD [OR: 4.50; 90% CI: (1.74-11.64); P = 0.009].
Table 2.
Association of HLA Alleles and Risk Factors with Acute Graft-Versus-Host Disease
|
Variables
|
aGVHD Grades 0-I (n=123)
|
aGVHD Grades II-IV (n=30)
|
Univariate
|
Multiplea
|
|
OR (75% CI)
|
P
value
*
|
AOR (90% CI)
|
P
value
**
|
| HLA-A29 |
|
| A29 + |
1 (33.3) |
2 (66.7) |
2.76 (1.35-5.66) |
0.098 |
|
|
| A29- (RL) |
88 (79.3) |
23 (20.7) |
1 |
|
|
| HLA-B07 |
|
| B07 + |
7 (53.8) |
6 (46.2) |
1.92 (1.35-2.73) |
0.031 |
3.84 (1.37-10.72) |
0.031 |
| B07- (RL) |
82 (81.2) |
19 (18.8) |
1 |
1 |
| HLA-B18 |
|
| B18 + |
7 (63.6) |
4 (36.4) |
1.49 (1.01-2.19) |
0.231 |
|
|
| B18 - (RL) |
82 (79.6) |
21 (20.4) |
1 |
|
|
| HLA-B51 |
|
| B51 + |
25 (89.3) |
3 (10.7) |
0.59 (0.40-0.86) |
0.108 |
|
|
| B51- (RL) |
64 (74.4) |
22 (25.6) |
1 |
|
|
| HLA-B52 |
|
| B52 + |
14 (93.3) |
1 (6.7) |
0.47 (0.25-0.86) |
0.152 |
|
|
| B52- (RL) |
75 (75.8) |
24 (24.2) |
1 |
|
|
| HLA-B57 |
|
| B57 + |
1 (33.3) |
2 (66.7) |
2.76 (1.35-5.66) |
0.103 |
|
|
| B57- (RL) |
88 (79.3) |
23 (20.7) |
1 |
|
|
| HLA-DRB1*07 |
|
| DRB1*07 + |
17 (68) |
8 (32) |
1.38 (1.03-1.85) |
0.198 |
2.11 (1.12-3.95) |
0.052 |
| DRB1*07- (RL) |
69 (80.2) |
17 (19.8) |
1 |
1 |
| HLA-DRB1*13 |
|
| DRB1*13 + |
18 (66.7) |
9 (33.3) |
1.45 (1.09-1.93) |
0.121 |
|
|
| DRB1*13- (RL) |
68 (81) |
16 (19) |
1 |
|
|
| HLA-DRB1*15 |
|
| DRB1*15 + |
28 (84.8) |
5 (15.2) |
0.72 (0.52-0.98) |
0.228 |
|
|
| DRB1*15- (RL) |
58 (74.4) |
20 (25.6) |
1 |
|
|
| Recipient CMV IgG |
|
| Positive |
56 (83.6) |
11 (16.4) |
1.07 (1.13-2.56) |
0.131 |
|
|
| Negative (RL) |
7 (63.6) |
4 (36.4) |
1 |
|
|
| Recipient gender |
|
| Female |
57 (76) |
18 (24) |
1.30 (1.03-1.65) |
0.194 |
|
|
| Male (RL) |
65 (84.4) |
12 (15.6) |
1 |
|
|
| Gender match |
|
| Female → Female |
22 (84.6) |
4 (15.4) |
0.79 (0.47-1.32) |
0.242 |
|
|
| Female → Male |
28 (77.8) |
8 (22.2) |
1.24 (0.82-1.88) |
0.589 |
|
|
| Male → Female |
33 (71.7) |
13 (28.3) |
1.71 (1.18-2.48) |
0.543 |
|
|
| Male → Male |
37 (30.8.4) |
5 (16.7) |
1 |
0.089 |
|
|
RL, reference level; AOR, adjusted odds ratio; aGVHD, acute graft-versus-host disease.
a Backward Selection; *Significant at 0.25; **Significant at 0.10.
Table 3.
Association of HLA Alleles and Risk Factors with Chronic Graft-Versus-Host Disease
|
Variables
|
cGVHD 0-Limited (n=122)
|
cGVHD Extensive (n=5)
|
Univariate
|
Multiplea
|
|
OR (75% CI)
|
P
value
*
|
OR (90% CI)
|
P
value
**
|
| HLA-A01 |
|
| A01 + |
15 (78.9) |
4 (21.1) |
4.44 (2.28-8.62) |
< 0.001 |
4.50 (1.74-11.64) |
0.009 |
| A01- (RL) |
74 (98.7) |
1 (1.3) |
1 |
1 |
| HLA-B07 |
|
| B07 + |
5 (83.3) |
1 (16.7) |
2.04 (1.02-4.10) |
0.228 |
|
|
| B07- (RL) |
84 (95.5) |
4 (4.5) |
1 |
|
|
| HLA-B40 |
|
| B40 + |
1 (50) |
1 (50) |
4.69 (1.97-11.13) |
0.029 |
|
|
| B40- (RL) |
88 (95.7) |
4 (4.3) |
1 |
|
|
| HLA-B41 |
|
| B41 + |
1 (50) |
1 (50) |
4.69 (1.97-11.13) |
0.029 |
|
|
| B41- (RL) |
88 (95.7) |
4 (4.3) |
1 |
|
|
| HLA-B57 |
|
| B57 + |
1 (50) |
1 (50) |
4.69 (1.97-11.13) |
0.029 |
|
|
| B57- (RL) |
88 (95.7) |
4 (4.3) |
1 |
|
|
| HLA-DRB1*08 |
|
| DRB1*08 + |
2 (66.7) |
1 (33.3) |
3.24 (1.51-6.95) |
0.072 |
|
|
| DRB1*08- (RL) |
84 (95.5) |
4 (4.5) |
1 |
|
|
| HLA-DRB1*16 |
|
| DRB1*16 + |
4 (80) |
1 (20) |
2.26 (1.11-4.59) |
0.178 |
|
|
| DRB1*16- (RL) |
82 (95.3) |
4 (4.7) |
1 |
|
|
RL, reference level; OR, Odds ratio; cGVHD, chronic graft-versus-host disease.
a Backward Selection; *Significant at 0.25; **Significant at 0.10.
The Effect of HLA Alleles on the Overall Survival
The influence of various HLA alleles on the OS are shown in Table 4. According to the Cox univariate model, 10 HLA alleles were significantly associated with mortality rate. The presence of A*66 in HLA-A was responsible for the greatest increase in the mortality rate (HR: 3.88; 75% CI: 1.19–12.63; P value = 0.182). Moreover, in HLA-B, B*40 and B*41 were accountable for the highest increase in the risk of death (HR: 4.71; 75% CI: 2.01–11.03; P value = 0.028). The presence of DRB1*02 in HLA-DRB1 caused the greatest increase in mortality rate (HR: 7.67; 75% CI: 2.30–25.50; P = 0.049). The patients with B*38 allele had a 72% decrease in the risk of mortality (HR: 0.28; 75% CI: 0.08–0.92; P = 0.221). The proportional hazards assumption based on the score process plot and the Kolmogorov-type supremum test was established for HLA-alleles in the final multivariable model and this model was valid. Regarding the final multivariable model, HLA-A*03, HLA-B*13, HLA-B*40, HLA-DRB1*02, and HLA-DRB1*04 were significantly linked to death rate and increased mortality risk in patients (Figure 2).
Table 4.
Association of HLA Alleles with Overall Survival
|
Variables
|
Univariate
|
Multiplea
|
|
HR(75% CI)
|
P
value
*
|
AHR(90% CI)
|
P
value
**
|
| HLA-A03 |
|
| A03 + |
1.99(1.36–2.93) |
0.029 |
1.88(1.01–3.52) |
0.089 |
| A03- (RL) |
1 |
1 |
| HLA-A31 |
|
| A31 + |
2.58(1.25–5.29) |
0.118 |
- |
- |
| A31- (RL) |
1 |
- |
| HLA-A66 |
|
| A66 + |
3.88(1.19–12.63) |
0.183 |
- |
- |
| A66- (RL) |
1 |
- |
| HLA-B07 |
|
| B07 + |
2.01 (1.27–3.19) |
0.068 |
- |
- |
| B07- (RL) |
1 |
- |
| HLA-B13 |
|
| B13 + |
2.16 (1.29–3.61) |
0.081 |
3.06 (1.39–6.72) |
0.013 |
| B13- (RL) |
1 |
1 |
| HLA-B38 |
|
| B38 + |
0.28 (0.08–0.92) |
0.221 |
- |
- |
| B38- (RL) |
1 |
- |
| HLA-B40 |
|
| B40 + |
4.71 (2.01–11.03) |
0.028 |
4.85 (1.35–17.36) |
0.042 |
| B40- (RL1) |
1 |
1 |
| HLA-B41 |
|
| B41 + |
4.71 (2.01–11.03) |
0.028 |
- |
- |
| B41- (RL) |
1 |
- |
| HLA-DRB1*02 |
|
| DRB1*02 + |
7.67 (2.30–25.50) |
0.049 |
7.12 (1.19–42.62) |
0.074 |
| DRB1*02- (RL) |
1 |
1 |
| HLA-DRB1*04 |
|
| DRB1*04 + |
2.17 (1.41–3.32) |
0.034 |
2.15 (1.13–4.10) |
0.039 |
| DRB1*04- (RL) |
1 |
1 |
RL, reference level; AHR, adjusted hazard ratio.
a Backward Selection; *Significant at 0.25; **Significant at 0.10.
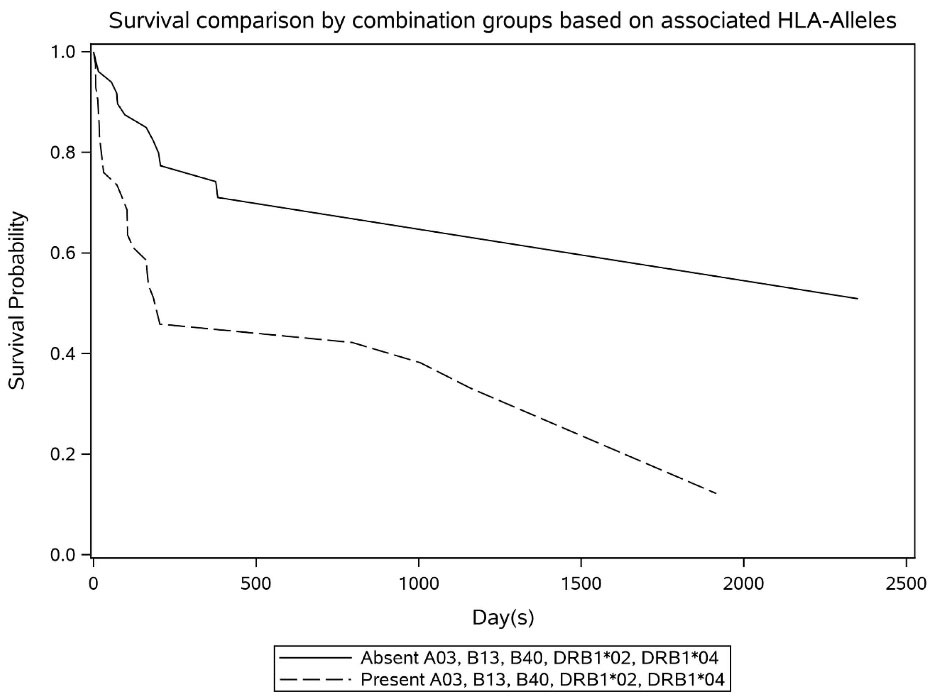
Figure 2.
Survival Trend Based on Important HLA-Allele Variants
.
Survival Trend Based on Important HLA-Allele Variants
Discussion
Donor-recipient HLA disparity, age and gender of both the recipient and the donor, multiparous female donors as well as graft source, GVHD prophylaxis regimens, and intensity of conditioning regimen are known as risk factors of GVHD occurrence.16 In this study, we focused on the HLA alleles frequency in the Iranian allo-HSCT patients and examined the association between the presence or absence of specific HLA alleles and incidence of acute and/or chronic GVHD in allo-HSCT patients with an HLA-matched sibling donor. We determined that acute GVHD grade II-IV occurrence was remarkably increased in the presence of HLA-B*07 and HLA-DRB1*07. In addition, it was observed that HLA-A*01 was related to a greater risk of chronic GVHD incidence. Furthermore, we analyzed the correlation of HLA alleles with OS. As exemplified in the results, the presence of any alleles of HLA-A*03, -B*13, B*40, -DRB1*02, or -DRB1*04 predicted a decline in the OS rate. Shawkatová and colleagues reported that patients who carry HLA-A*01, -DRB1*03 and -DQB1*03 alleles had lower incidence of aGVHD. In contrast, the HLA-DQB1*06 allele increased the incidence of cGVHD.17
Identification of the specific HLA alleles, associated with increasing or decreasing incidence ofacute or chronic GVHD and OS may have specific clinical implications in each population. Hence, the association of HLA loci or haplotypes with allo-HSCT outcomes have been evaluated in different populations. Storbet al estimated that the presence of HLA-B*18 is associated with a nearly three-fold increase in aGVHD incidence, whereas in patients with HLA-B*8 or HLA-Bw35, the incidence of aGVHD decreased by half, in comparison with those without these alleles.18 The European group for Blood and Marrow Transplantation (EBMT) conducted a cohort study in CML patients who received transplantation from HLA-matched sibling donors. In this homogenous large study, HLA-A*3 and HLA-DR1 significantly raised and diminished the occurrence of aGVHD grade II-IV, respectively.19 As our results suggested, HLA-B*7 was the HLA allele with the highest risk of aGVHD grade II-IV. Remberger et al20 pointed out that the HLA alleles A*10 and B*7 had a significant effect on enhancing the risk of acute GVHD grades II-IV. The chronic GVHD incidence was also low in patients who had the HLA-B27 allele. Ghavamzadeh et al reported an increased risk of severe acute grade III and IV GVHD in patients with HLA-DRB1*07. Our result confirmed this association in a way that these alleles could increase the risk of aGVHD grades II-IV by more than two times. Moreover, they suggested a protective role against severe a-GVHD for HLA-B35 in 162 recipients of HLA-identical sibling donor with different diseases.21 Given that the population ethnicity and sample size was similar to our study, some parallel/contradictory findings need further confirmation. Separate studies by the same center confirmed that the presence of HLA-A*11 and HLA-A*26 increased the risk of aGVHD, whereas HLA-A*3 reduced its risk in thalassemic patients who underwent HLA-Identical HSCT.22 A single-center experience in Korea demonstrated that the incidence of grades II to IV aGVHD was higher in patients with HLA-B*61 and HLA-Cw3. HLA-B*54 had close relationship with a higher incidence of extensive-type cGVHD. Furthermore, evaluating the association between HLA alleles and organ-related GVHD showed HLA-B*35, HLA-B*54, and HLA-B*7, B*27 to be associated with the development of severe acute skin GVHD, chronic skin/oral GVHD, and chronic liver GVHD, respectively. They suggested that these remarkable HLA alleles may be potent transplantation immune regulators.8 In a Brazilian population, increased incidence of acute GVHD grade III or higher was positively correlated with HLA-B*35, B*49, B*55 and in patients with extensive chronic GVHD, HLA-A*9, A*24 and A*26 were higher than other patients, while HLA-A*2 was lower.9
Racial difference is one of the main factors that lead to the discrepancy between various HLA alleles linked to the risk of GVHD in different studies. In the context of HLA-matched transplantation, the role of specific HLA alleles in decreasing or increasing the incidence of GVHD could be explained by different mechanisms. One possible mechanism is the nature of the HLA system function in determining immune responses. As we know, the specificity of HLA–peptide–TCR interactions is fundamental in the function of the adaptive immune system.23 HLA molecules play a substantial role in mounting an efficient and appropriate response to non-self antigens and maintaining self-tolerance to prevent autoimmune diseases.24 Therefore, the presence or absence of various HLA molecules might determine responsiveness and/or unresponsiveness of the immune system against foreign antigens. For instance, HLADRB1*15 is known as a marker of clinical response to immunosuppressive therapy in autoimmune cytopenia.25 Battiwalla et al. reported that the presence of HLA-DRB1*15 in patients with myeloid malignancies was associated with a significantly lower incidence of aGVHD. Accordingly, we found a decreased incidence of aGVHD in HLA-DRB1*15 positive patients. There is a hypothesis that HLA-DRB1*15 could be a marker for a generalized susceptibility to immunosuppression.26
Another possible mechanism for explicating GVHD occurrence in patients who receive HLA-matched transplantation is donor-recipient minor histocompatibility antigens (MiHAs) disparity. MiHAs are polymorphic peptide fragments which are derived from intracellular proteins. Despite the fact that MiHAs inheritance is independent of HLA molecules, they are presented by MHC class I and II and can be recognized by alloreactive T cells leading to induction of immune responses.27 It has been opined that various HLA alleles may have different capabilities to present MiHA that are responsible for GVHD in HLA-matched transplantation. A study reported some minor H and HY antigens which can induce T-cell responses more often than the others.28 Some of these H antigens (HA-1 and HA-2) are HLA-A*0201-restricted antigens which are solely expressed in hematopoietic-originated cells. Mismatching of HA-1 between donor-recipient in the context of HLA-matched bone marrow transplant was significantly associated with acute GVHD.29 Recently, MiHAs-based immunotherapy strategies have been considered as an interesting candidate not only to prevent GVHD development but also for inducing an effective GVT response.30
On the other hand, some inflammatory mediators including histamine, prostaglandin, corticosteroids and cytokines, especially tumor necrosis factor alpha (TNF-α) which are pivotal in the pathophysiology of GVHD are encoded by HLA-related genes.31 Bouma et al demonstrated that the HLA-DR1-related TNF haplotype presents a low-secretor phenotype and produces lower levels of TNF-α in comparison with other HLA alleles. Consequently, reducing TNF-α secretion might provoke the HLA-DR1 protective effect on GVHD occurrence.32
In conclusion, our data suggests that HLA alleles have independent influential effects on the occurrence of GVHD and also in OS of patients undergoing allo-HSCT, even in patients receiving the graft from an HLA-matched sibling donor. This might allude that certain HLA-antigens could act as an immunogenic stimulus to prompt the occurrence of GVHD and probably change the survival rate of patients. Furthermore, it is possible that MiHAs which are restricted to specific HLA alleles contribute to the incidence and severity of GVHD and HSCT outcome. Finding the contributing HLA alleles to the increased or decreased risk of GVHD and mortality might assist in selecting the strategies for prevention/treatment of GVHD and improve survival rate in HLA-matched HSCT patients. Our study was limited by the relatively small number of patients, absence of HLA-C, DR, and DQ typing for all patients who underwent HLA-matched sibling allo-HSCT between 2009 and 2018 at Taleghani hospital, and a the single-center nature of the study. Confirmation of the findings needs larger multi-center studies encompassing more HLA alleles and even MiHAs.
Acknowledgements
The authors would like to thank the staff of the Hematopoietic Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, whose support and collaboration made this study possible.
Competing Interests
All authors declare that they have no conflict of interest.
Ethical Approval
The study was approved by the ethical committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran (IR.SBMU.RETECH.REC.1399.484). Informed consents were obtained from all patients. Retrospective studies are conducted on already available data, and all the procedures being performed were part of the routine care.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Gyurkocza B, Rezvani A, Storb RF. Allogeneic hematopoietic cell transplantation: the state of the art. Expert Rev Hematol 2010; 3(3):285-99. doi: 10.1586/ehm.10.21 [Crossref] [ Google Scholar]
- Ghimire S, Weber D, Mavin E, Wang XN, Dickinson AM, Holler E. Pathophysiology of GvHD and other HSCT-related major complications. Front Immunol 2017; 8:79. doi: 10.3389/fimmu.2017.00079 [Crossref] [ Google Scholar]
- Couriel D, Caldera H, Champlin R, Komanduri K. Acute graft-versus-host disease: pathophysiology, clinical manifestations, and management. Cancer 2004; 101(9):1936-46. doi: 10.1002/cncr.20613 [Crossref] [ Google Scholar]
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 2009; 54(1):15-39. doi: 10.1038/jhg.2008.5 [Crossref] [ Google Scholar]
- Claas FH, Duquesnoy RJ. The polymorphic alloimmune response in clinical transplantation. Curr Opin Immunol 2008; 20(5):566-7. doi: 10.1016/j.coi.2008.08.001 [Crossref] [ Google Scholar]
- Petersdorf EW. Which factors influence the development of GVHD in HLA-matched or mismatched transplants?. Best Pract Res Clin Haematol 2017; 30(4):333-5. doi: 10.1016/j.beha.2017.09.003 [Crossref] [ Google Scholar]
- Kuba A, Raida L. Graft versus host disease: from basic pathogenic principles to DNA damage response and cellular senescence. Mediators Inflamm 2018; 2018:9451950. doi: 10.1155/2018/9451950 [Crossref] [ Google Scholar]
- Kim HJ, Park SJ, Im HW, Kim DW, Min WS, Kim HK. The association of HLA antigen and GVHD in allogeneic hemopoietic stem cell transplantation with histocompatible sibling donor: a single-center experience in Korea. Int J Hematol 2002; 76(3):267-71. doi: 10.1007/bf02982797 [Crossref] [ Google Scholar]
- Cardozo DM, Lieber SR, Marques SB, Aranha FJ, Vigorito AC, de Souza CA. Association between human leukocyte antigens and graft-versus-host disease occurrence after allogenic hematopoietic stem cell transplantation. Sao Paulo Med J 2012; 130(4):219-24. doi: 10.1590/s1516-31802012000400003 [Crossref] [ Google Scholar]
- Martin PJ, Gooley T, Anasetti C, Petersdorf EW, Hansen JA. HLAs and risk of acute graft-vs-host disease after marrow transplantation from an HLA-identical sibling. Biol Blood Marrow Transplant 1998; 4(3):128-33. doi: 10.1053/bbmt.1998.v4.pm9923410 [Crossref] [ Google Scholar]
- Parkhideh S, Chegeni R, Mehdizadeh M, Roshandel E, Tavakoli F, Hajifathali A. Effects of ABO incompatibility on the outcome of allogeneic hematopoietic stem cell transplantation. Transfus Apher Sci 2020; 59(2):102696. doi: 10.1016/j.transci.2019.102696 [Crossref] [ Google Scholar]
- Ghasemi K, Parkhideh S, Kazemi MH, Salimi M, Salari S, Nalini R. The role of serum uric acid in the prediction of graft-versus-host disease in allogeneic hematopoietic stem cell transplantation. J Clin Lab Anal 2020; 34(7):e23271. doi: 10.1002/jcla.23271 [Crossref] [ Google Scholar]
- Tavakoli Ardakani M, Mehrpooya M, Mehdizadeh M, Beiraghi N, Hajifathali A, Kazemi MH. Sertraline treatment decreased the serum levels of interleukin-6 and high-sensitivity C-reactive protein in hematopoietic stem cell transplantation patients with depression; a randomized double-blind, placebo-controlled clinical trial. Bone Marrow Transplant 2020; 55(4):830-2. doi: 10.1038/s41409-019-0623-0 [Crossref] [ Google Scholar]
- Dignan FL, Clark A, Amrolia P, Cornish J, Jackson G, Mahendra P. Diagnosis and management of acute graft-versus-host disease. Br J Haematol 2012; 158(1):30-45. doi: 10.1111/j.1365-2141.2012.09129.x [Crossref] [ Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16(3):1215. doi: 10.1093/nar/16.3.1215 [Crossref] [ Google Scholar]
- Nassereddine S, Rafei H, Elbahesh E, Tabbara I. Acute graft versus host disease: a comprehensive review. Anticancer Res 2017; 37(4):1547-55. doi: 10.21873/anticanres.11483 [Crossref] [ Google Scholar]
- Shawkatová I, Bojtárová E, Kováčová M, Klučková K, Kušíková M, Mistrík M. Individual HLA alleles and risk of graft-versus-host disease after haematopoietic stem cell transplantation from HLA-identical siblings. Biologia 2020; 75(11):2045-52. doi: 10.2478/s11756-020-00510-1 [Crossref] [ Google Scholar]
- Storb R, Prentice RL, Hansen JA, Thomas ED. Association between HLA-B antigens and acute graft-versus-host disease. Lancet 1983; 2(8354):816-9. doi: 10.1016/s0140-6736(83)90737-7 [Crossref] [ Google Scholar]
- Clark RE, Hermans J, Madrigal A, Nachbaur D, Kropshofer G, Gratwohl A. HLA-A3 increases and HLA-DR1 decreases the risk of acute graft-versus-host disease after HLA-matched sibling bone marrow transplantation for chronic myelogenous leukaemia. Br J Haematol 2001; 114(1):36-41. doi: 10.1046/j.1365-2141.2001.02897.x [Crossref] [ Google Scholar]
- Remberger M, Persson U, Hauzenberger D, Ringdén O. An association between human leucocyte antigen alleles and acute and chronic graft-versus-host disease after allogeneic haematopoietic stem cell transplantation. Br J Haematol 2002; 119(3):751-9. doi: 10.1046/j.1365-2141.2002.03924.x [Crossref] [ Google Scholar]
- Ghavamzadeh A, Alimoghaddam K, Behrouzan O, Bahar B, Jahani M, Sajadi SA. HLA and risk of acute graft versus host disease in allogeneic bone marrow transplantation from an HLA-identical sibling. Arch Iran Med 2002; 5(1):16-20. [ Google Scholar]
- Mohyeddin Bonab M, Alimoghaddam K, Vatandoust S, Forouzia F, Jahani M, Ghavamzadeh A. Are HLA antigens a risk factor for acute GVHD in thalassemic patients receiving HLA-identical stem cell transplantation?. Transplant Proc 2004; 36(10):3190-3. doi: 10.1016/j.transproceed.2004.10.088 [Crossref] [ Google Scholar]
- Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol 2018; 18(5):325-39. doi: 10.1038/nri.2017.143 [Crossref] [ Google Scholar]
- Shankarkumar U. The human leukocyte antigen (HLA) system. Int J Hum Genet 2004; 4(2):91-103. doi: 10.1080/09723757.2004.11885875 [Crossref] [ Google Scholar]
- Saunthararajah Y, Nakamura R, Nam JM, Robyn J, Loberiza F, Maciejewski JP. HLA-DR15 (DR2) is overrepresented in myelodysplastic syndrome and aplastic anemia and predicts a response to immunosuppression in myelodysplastic syndrome. Blood 2002; 100(5):1570-4. [ Google Scholar]
- Battiwalla M, Hahn T, Radovic M, Roy H, Wahab A, Duman E. Human leukocyte antigen (HLA) DR15 is associated with reduced incidence of acute GVHD in HLA-matched allogeneic transplantation but does not impact chronic GVHD incidence. Blood 2006; 107(5):1970-3. doi: 10.1182/blood-2005-05-1958 [Crossref] [ Google Scholar]
- Roy DC, Perreault C. Major vs minor histocompatibility antigens. Blood 2017; 129(6):664-6. doi: 10.1182/blood-2016-12-754515 [Crossref] [ Google Scholar]
- Hobo W, Broen K, van der Velden WJ, Greupink-Draaisma A, Adisty N, Wouters Y. Association of disparities in known minor histocompatibility antigens with relapse-free survival and graft-versus-host disease after allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2013; 19(2):274-82. doi: 10.1016/j.bbmt.2012.09.008 [Crossref] [ Google Scholar]
- Goulmy E, Schipper R, Pool J, Blokland E, Falkenburg JH, Vossen J. Mismatches of minor histocompatibility antigens between HLA-identical donors and recipients and the development of graft-versus-host disease after bone marrow transplantation. N Engl J Med 1996; 334(5):281-5. doi: 10.1056/nejm199602013340501 [Crossref] [ Google Scholar]
- Franssen LE, Roeven MWH, Hobo W, Doorn R, Oostvogels R, Falkenburg JHF. A phase I/II minor histocompatibility antigen-loaded dendritic cell vaccination trial to safely improve the efficacy of donor lymphocyte infusions in myeloma. Bone Marrow Transplant 2017; 52(10):1378-83. doi: 10.1038/bmt.2017.118 [Crossref] [ Google Scholar]
- Takahashi H, Furukawa T, Hashimoto S, Suzuki N, Kuroha T, Yamazaki F. Contribution of TNF-alpha and IL-10 gene polymorphisms to graft-versus-host disease following allo-hematopoietic stem cell transplantation. Bone Marrow Transplant 2000; 26(12):1317-23. doi: 10.1038/sj.bmt.1702724 [Crossref] [ Google Scholar]
- Bouma G, Crusius JB, Oudkerk Pool M, Kolkman JJ, von Blomberg BM, Kostense PJ. Secretion of tumour necrosis factor alpha and lymphotoxin alpha in relation to polymorphisms in the TNF genes and HLA-DR alleles Relevance for inflammatory bowel disease. Scand J Immunol 1996; 43(4):456-63. doi: 10.1046/j.1365-3083.1996.d01-65.x [Crossref] [ Google Scholar]